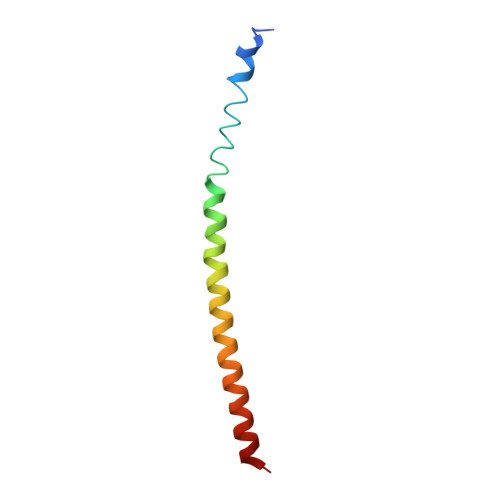
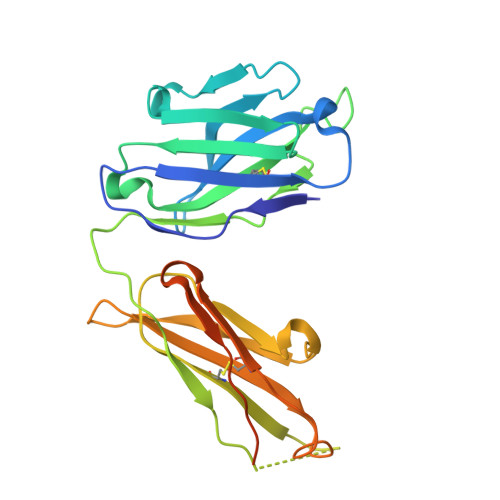
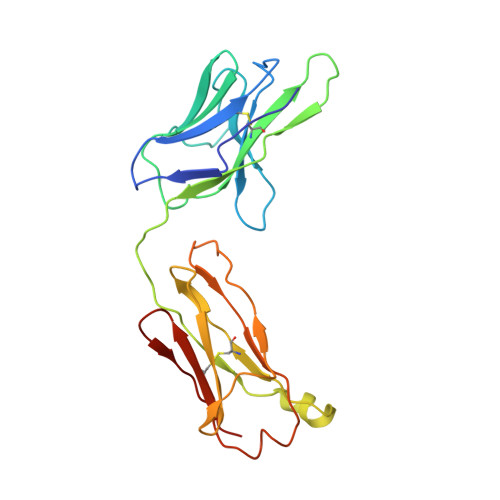
Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41.
Gustchina, E., Li, M., Ghirlando, R., Schuck, P., Louis, J.M., Pierson, J., Rao, P., Subramaniam, S., Gustchina, A., Clore, G.M., Wlodawer, A.(2013) PLoS One 8: e78187-e78187
- PubMed: 24244293
- DOI: https://doi.org/10.1371/journal.pone.0078187
- Primary Citation of Related Structures:
4KHT, 4KHX - PubMed Abstract:
A series of mini-antibodies (monovalent and bivalent Fabs) targeting the conserved internal trimeric coiled-coil of the N-heptad repeat (N-HR) of HIV-1 gp41 has been previously constructed and reported. Crystal structures of two closely related monovalent Fabs, one (Fab 8066) broadly neutralizing across a wide panel of HIV-1 subtype B and C viruses, and the other (Fab 8062) non-neutralizing, representing the extremes of this series, were previously solved as complexes with 5-Helix, a gp41 pre-hairpin intermediate mimetic. Binding of these Fabs to covalently stabilized chimeric trimers of N-peptides of HIV-1 gp41 (named (CCIZN36)3 or 3-H) has now been investigated using X-ray crystallography, cryo-electron microscopy, and a variety of biophysical methods. Crystal structures of the complexes between 3-H and Fab 8066 and Fab 8062 were determined at 2.8 and 3.0 Å resolution, respectively. Although the structures of the complexes with the neutralizing Fab 8066 and its non-neutralizing counterpart Fab 8062 were generally similar, small differences between them could be correlated with the biological properties of these antibodies. The conformations of the corresponding CDRs of each antibody in the complexes with 3-H and 5-Helix are very similar. The adaptation to a different target upon complex formation is predominantly achieved by changes in the structure of the trimer of N-HR helices, as well as by adjustment of the orientation of the Fab molecule relative to the N-HR in the complex, via rigid-body movement. The structural data presented here indicate that binding of three Fabs 8062 with high affinity requires more significant changes in the structure of the N-HR trimer compared to binding of Fab 8066. A comparative analysis of the structures of Fabs complexed to different gp41 intermediate mimetics allows further evaluation of biological relevance for generation of neutralizing antibodies, as well as provides novel structural insights into immunogen design.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland, United States of America.
Organizational Affiliation: