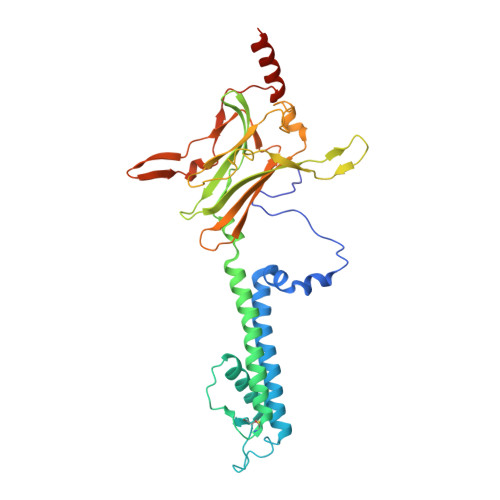
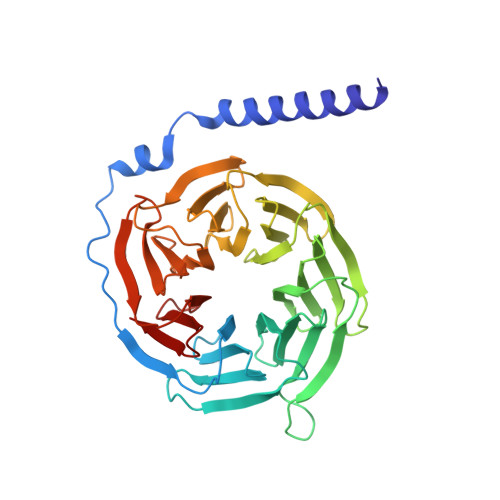
X-ray structure of the mammalian GIRK2-beta gamma G-protein complex.
Whorton, M.R., MacKinnon, R.(2013) Nature 498: 190-197
- PubMed: 23739333
- DOI: https://doi.org/10.1038/nature12241
- Primary Citation of Related Structures:
4KFM - PubMed Abstract:
G-protein-gated inward rectifier K(+) (GIRK) channels allow neurotransmitters, through G-protein-coupled receptor stimulation, to control cellular electrical excitability. In cardiac and neuronal cells this control regulates heart rate and neural circuit activity, respectively. Here we present the 3.5 Å resolution crystal structure of the mammalian GIRK2 channel in complex with βγ G-protein subunits, the central signalling complex that links G-protein-coupled receptor stimulation to K(+) channel activity. Short-range atomic and long-range electrostatic interactions stabilize four βγ G-protein subunits at the interfaces between four K(+) channel subunits, inducing a pre-open state of the channel. The pre-open state exhibits a conformation that is intermediate between the closed conformation and the open conformation of the constitutively active mutant. The resultant structural picture is compatible with 'membrane delimited' activation of GIRK channels by G proteins and the characteristic burst kinetics of channel gating. The structures also permit a conceptual understanding of how the signalling lipid phosphatidylinositol-4,5-bisphosphate (PIP2) and intracellular Na(+) ions participate in multi-ligand regulation of GIRK channels.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, 1230 York Avenue, New York, New York 10065, USA.
Organizational Affiliation: