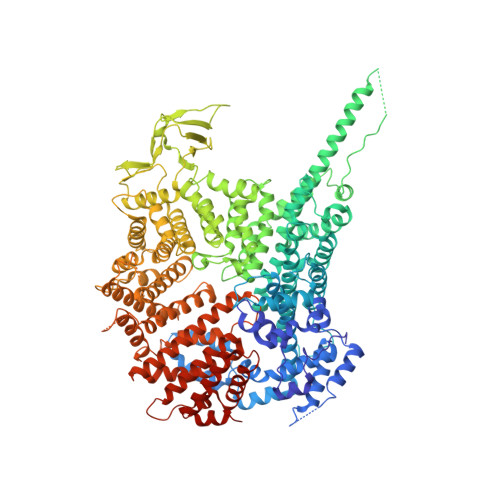
Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors.
Andersen, K.R., Onischenko, E., Tang, J.H., Kumar, P., Chen, J.Z., Ulrich, A., Liphardt, J.T., Weis, K., Schwartz, T.U.(2013) Elife 2: e00745-e00745
- PubMed: 23795296
- DOI: https://doi.org/10.7554/eLife.00745
- Primary Citation of Related Structures:
4KF7, 4KF8 - PubMed Abstract:
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs) embedded in the nuclear envelope. About 30 different proteins (nucleoporins, nups) arrange around a central eightfold rotational axis to build the modular NPC. Nup188 and Nup192 are related and evolutionary conserved, large nucleoporins that are part of the NPC scaffold. Here we determine the structure of Nup188. The protein folds into an extended stack of helices where an N-terminal 130 kDa segment forms an intricate closed ring, while the C-terminal region is a more regular, superhelical structure. Overall, the structure has distant similarity with flexible S-shaped nuclear transport receptors (NTRs). Intriguingly, like NTRs, both Nup188 and Nup192 specifically bind FG-repeats and are able to translocate through NPCs by facilitated diffusion. This blurs the existing dogma of a clear distinction between stationary nups and soluble NTRs and suggests an evolutionary relationship between the NPC and the soluble nuclear transport machinery. DOI:http://dx.doi.org/10.7554/eLife.00745.001.
- Department of Biology , Massachusetts Institute of Technology , Cambridge , United States.
Organizational Affiliation: