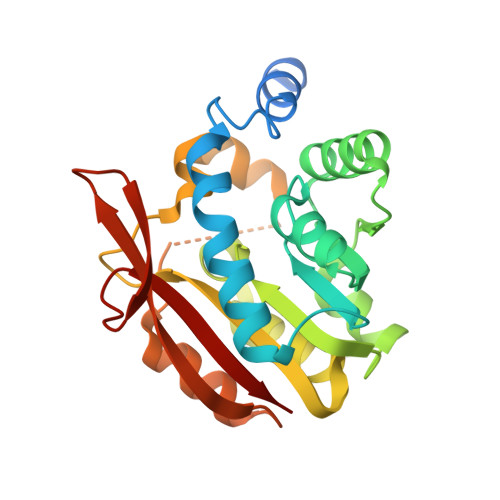
Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Zhu, Y., Wu, B., Zhang, X., Fan, X., Niu, L., Li, X., Wang, J., Teng, M.(2015) Biochem J 470: 105-114
- PubMed: 26251450
- DOI: https://doi.org/10.1042/BJ20150329
- Primary Citation of Related Structures:
4KDC - PubMed Abstract:
UbiG and Coq3 (orthologue in eukaryotes) are SAM-MTases (S-adenosylmethionine-dependent methyltransferases) that catalyse both O-methylation steps in CoQ biosynthesis from prokaryotes to eukaryotes. However, the detailed molecular mechanism by which they function remains elusive. In the present paper, we report that UbiG/Coq3 defines a novel class of membrane-binding proteins. Escherichia coli UbiG binds specifically to liposomes containing PG (phosphatidylglycerol) or CL (cardiolipin, or diphosphatidylglycerol), two major lipid components of the E. coli plasma membrane, whereas human and yeast Coq3 display a strong preference for liposomes enriched with CL, a signature lipid of the mitochondrial membrane. The crystal structure of UbiG from E. coli was determined at 2.1 Å (1 Å = 0.1 nm) resolution. The structure exhibits a typical Class I SAM-MTase fold with several variations, including a unique insertion between strand β5 and helix α10. This insertion is highly conserved and is required for membrane binding. Mutation of the key residues renders UbiG unable to efficiently bind liposome in vitro and the mutant fails to rescue the phenotype of ΔubiG strain in vivo. Taken together, our results shed light on a novel biochemical function of the UbiG/Coq3 protein.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China.
Organizational Affiliation: