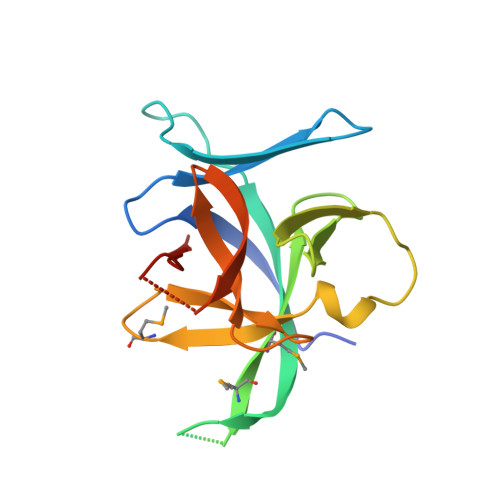
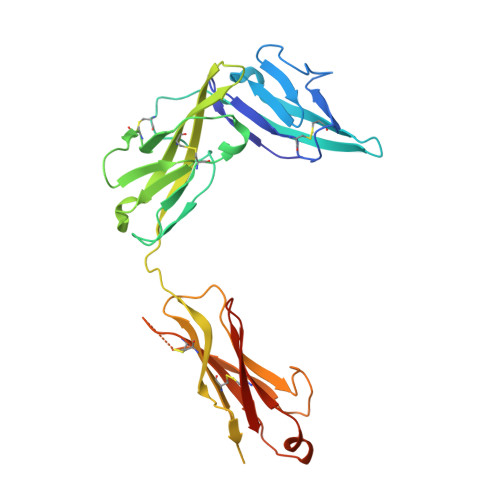
Structural insights into the interaction of IL-33 with its receptors.
Liu, X., Hammel, M., He, Y., Tainer, J.A., Jeng, U.S., Zhang, L., Wang, S., Wang, X.(2013) Proc Natl Acad Sci U S A 110: 14918-14923
- PubMed: 23980170
- DOI: https://doi.org/10.1073/pnas.1308651110
- Primary Citation of Related Structures:
4KC3 - PubMed Abstract:
Interleukin (IL)-33 is an important member of the IL-1 family that has pleiotropic activities in innate and adaptive immune responses in host defense and disease. It signals through its ligand-binding primary receptor ST2 and IL-1 receptor accessory protein (IL-1RAcP), both of which are members of the IL-1 receptor family. To clarify the interaction of IL-33 with its receptors, we determined the crystal structure of IL-33 in complex with the ectodomain of ST2 at a resolution of 3.27 Å. Coupled with structure-based mutagenesis and binding assay, the structural results define the molecular mechanism by which ST2 specifically recognizes IL-33. Structural comparison with other ligand-receptor complexes in the IL-1 family indicates that surface-charge complementarity is critical in determining ligand-binding specificity of IL-1 primary receptors. Combined crystallography and small-angle X-ray-scattering studies reveal that ST2 possesses hinge flexibility between the D3 domain and D1D2 module, whereas IL-1RAcP exhibits a rigid conformation in the unbound state in solution. The molecular flexibility of ST2 provides structural insights into domain-level conformational change of IL-1 primary receptors upon ligand binding, and the rigidity of IL-1RAcP explains its inability to bind ligands directly. The solution architecture of IL-33-ST2-IL-1RAcP complex from small-angle X-ray-scattering analysis resembles IL-1β-IL-1RII-IL-1RAcP and IL-1β-IL-1RI-IL-1RAcP crystal structures. The collective results confer IL-33 structure-function relationships, supporting and extending a general model for ligand-receptor assembly and activation in the IL-1 family.
- Ministry of Education Key Laboratory of Protein Science, Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: