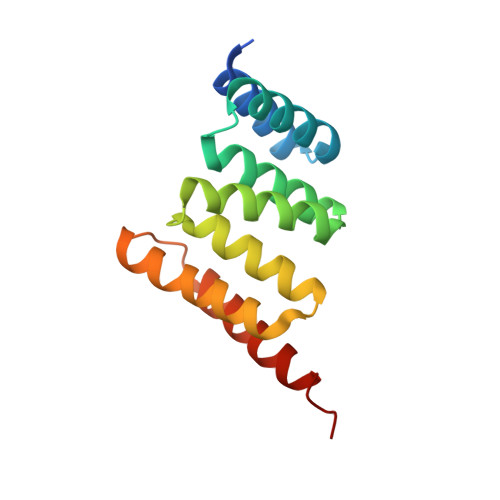
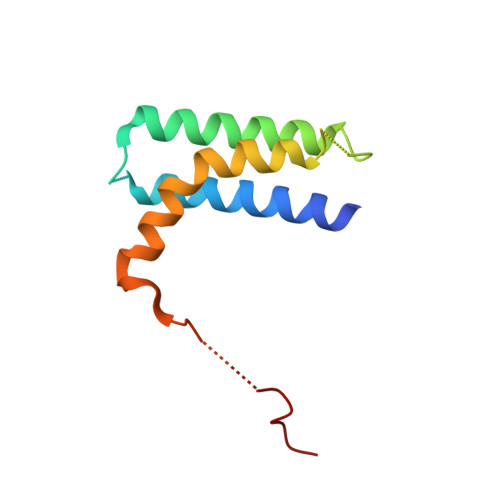
A Bipartite Interaction between Hsp70 and CHIP Regulates Ubiquitination of Chaperoned Client Proteins.
Zhang, H., Amick, J., Chakravarti, R., Santarriaga, S., Schlanger, S., McGlone, C., Dare, M., Nix, J.C., Scaglione, K.M., Stuehr, D.J., Misra, S., Page, R.C.(2015) Structure 23: 472-482
- PubMed: 25684577
- DOI: https://doi.org/10.1016/j.str.2015.01.003
- Primary Citation of Related Structures:
4KBQ - PubMed Abstract:
The ubiquitin ligase CHIP plays an important role in cytosolic protein quality control by ubiquitinating proteins chaperoned by Hsp70/Hsc70 and Hsp90, thereby targeting such substrate proteins for degradation. We present a 2.91 Å resolution structure of the tetratricopeptide repeat (TPR) domain of CHIP in complex with the α-helical lid subdomain and unstructured tail of Hsc70. Surprisingly, the CHIP-TPR interacts with determinants within both the Hsc70-lid subdomain and the C-terminal PTIEEVD motif of the tail, exhibiting an atypical mode of interaction between chaperones and TPR domains. We demonstrate that the interaction between CHIP and the Hsc70-lid subdomain is required for proper ubiquitination of Hsp70/Hsc70 or Hsp70/Hsc70-bound substrate proteins. Posttranslational modifications of the Hsc70 lid and tail disrupt key contacts with the CHIP-TPR and may regulate CHIP-mediated ubiquitination. Our study shows how CHIP docks onto Hsp70/Hsc70 and defines a bipartite mode of interaction between TPR domains and their binding partners.
- Department of Chemistry and Biochemistry, Miami University, Oxford, OH 45056, USA.
Organizational Affiliation: