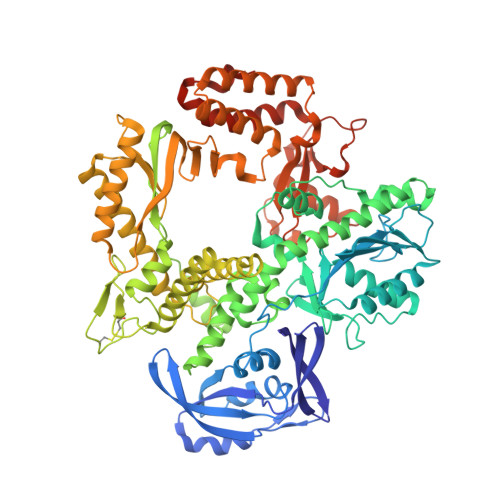
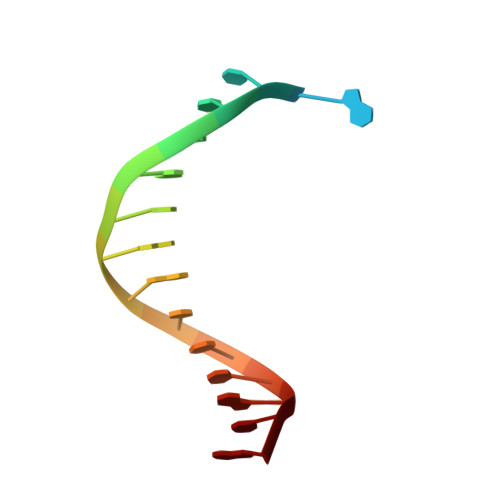
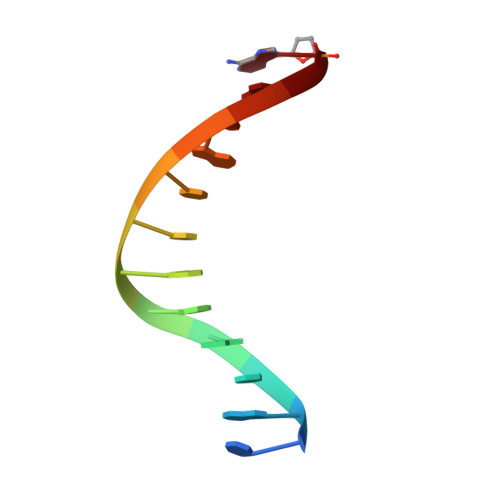
Structures of KOD and 9N DNA Polymerases Complexed with Primer Template Duplex
Bergen, K., Betz, K., Welte, W., Diederichs, K., Marx, A.(2013) Chembiochem 14: 1058-1062
- PubMed: 23733496
- DOI: https://doi.org/10.1002/cbic.201300175
- Primary Citation of Related Structures:
4K8X, 4K8Z - PubMed Abstract:
Replicate it: Structures of KOD and 9°N DNA polymerases, two enzymes that are widely used to replicate DNA with highly modified nucleotides, were solved at high resolution in complex with primer/template duplex. The data elucidate substrate interaction of the two enzymes and pave the way for further optimisation of the enzymes and substrates.
- Department of Chemistry, Konstanz Research School Chemical Biology, University of Konstanz, Universitätsstrasse 10, 78457 Konstanz, Germany.
Organizational Affiliation: