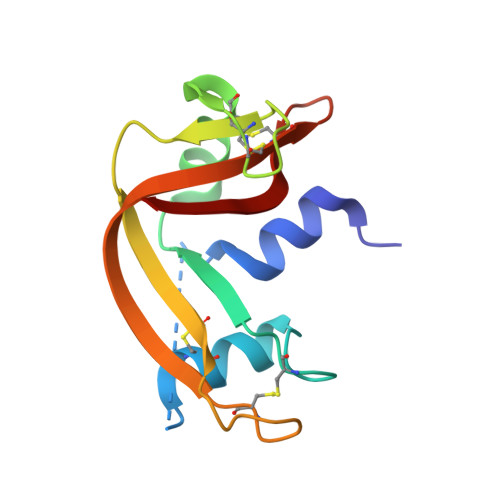
Crystal Structure of Apo- and Metalated Thiolate containing RNase S as Structural Basis for the Design of Artificial Metalloenzymes by Peptide- Protein Complementation
Genz, M., Singer, D., Hey-Hawkins, E., Hoffmann, R., Straeter, N.(2013) Z Anorg Allg Chem 639: 2395-2400