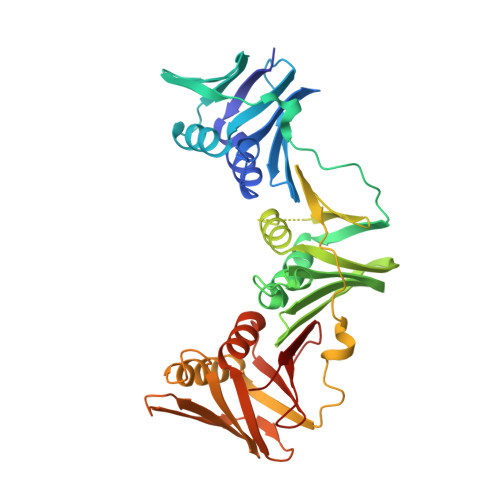
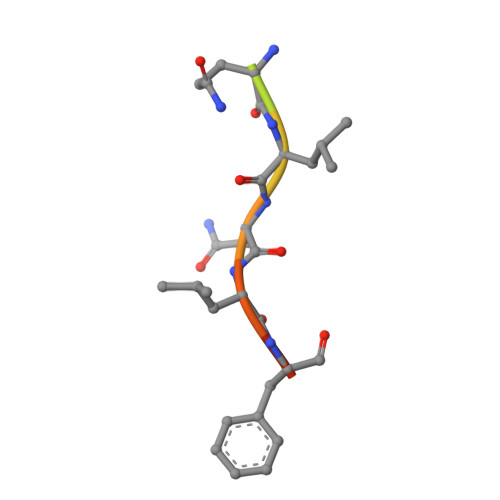
The UmuC subunit of the E. coli DNA polymerase V shows a unique interaction with the beta-clamp processivity factor.
Patoli, A.A., Winter, J.A., Bunting, K.A.(2013) BMC Struct Biol 13: 12-12
- PubMed: 23822808
- DOI: https://doi.org/10.1186/1472-6807-13-12
- Primary Citation of Related Structures:
4K74 - PubMed Abstract:
Strict regulation of replisome components is essential to ensure the accurate transmission of the genome to the next generation. The sliding clamp processivity factors play a central role in this regulation, interacting with both DNA polymerases and multiple DNA processing and repair proteins. Clamp binding partners share a common peptide binding motif, the nature of which is essentially conserved from phage through to humans. Given the degree of conservation of these motifs, much research effort has focussed on understanding how the temporal and spatial regulation of multiple clamp binding partners is managed. The bacterial sliding clamps have come under scrutiny as potential targets for rational drug design and comprehensive understanding of the structural basis of their interactions is crucial for success. In this study we describe the crystal structure of a complex of the E. coli β-clamp with a 12-mer peptide from the UmuC protein. UmuC is the catalytic subunit of the translesion DNA polymerase, Pol V (UmuD'₂C). Due to its potentially mutagenic action, Pol V is tightly regulated in the cell to limit access to the replication fork. Atypically for the translesion polymerases, both bacterial and eukaryotic, Pol V is heterotrimeric and its β-clamp binding motif (³⁵⁷QLNLF³⁶¹) is internal to the protein, rather than at the more usual C-terminal position. Our structure shows that the UmuC peptide follows the overall disposition of previously characterised structures with respect to the highly conserved glutamine residue. Despite good agreement with the consensus β-clamp binding motif, distinct variation is shown within the hydrophobic binding pocket. While UmuC Leu-360 interacts as noted in other structures, Phe-361 does not penetrate the pocket at all, sitting above the surface. Although the β-clamp binding motif of UmuC conforms to the consensus sequence, variation in its mode of clamp binding is observed compared to related structures, presumably dictated by the proximal aspartate residues that act as linker to the poorly characterised, unique C-terminal domain of UmuC. Additionally, interactions between Asn-359 of UmuC and Arg-152 on the clamp surface may compensate for the reduced interaction of Phe-361.
- Centre for Genetics and Genomics, University of Nottingham, Queen's Medical Centre, Nottingham NG7 2UH, UK.
Organizational Affiliation: