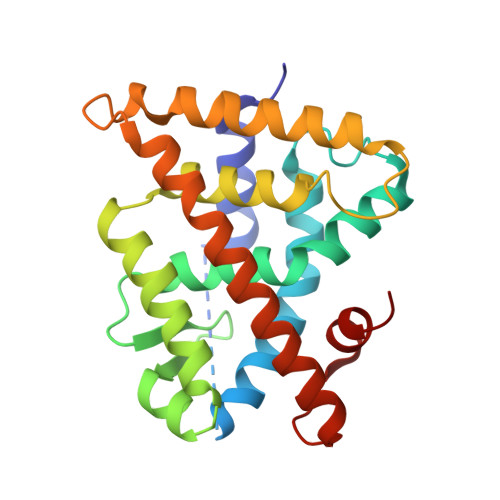
Defining the Communication between Agonist and Coactivator Binding in the Retinoid X Receptor alpha Ligand Binding Domain.
Boerma, L.J., Xia, G., Qui, C., Cox, B.D., Chalmers, M.J., Smith, C.D., Lobo-Ruppert, S., Griffin, P.R., Muccio, D.D., Renfrow, M.B.(2014) J Biological Chem 289: 814-826
- PubMed: 24187139
- DOI: https://doi.org/10.1074/jbc.M113.476861
- Primary Citation of Related Structures:
4K4J, 4K6I - PubMed Abstract:
Retinoid X receptors (RXRs) are obligate partners for several other nuclear receptors, and they play a key role in several signaling processes. Despite being a promiscuous heterodimer partner, this nuclear receptor is a target of therapeutic intervention through activation using selective RXR agonists (rexinoids). Agonist binding to RXR initiates a large conformational change in the receptor that allows for coactivator recruitment to its surface and enhanced transcription. Here we reveal the structural and dynamical changes produced when a coactivator peptide binds to the human RXRα ligand binding domain containing two clinically relevant rexinoids, Targretin and 9-cis-UAB30. Our results show that the structural changes are very similar for each rexinoid and similar to those for the pan-agonist 9-cis-retinoic acid. The four structural changes involve key residues on helix 3, helix 4, and helix 11 that move from a solvent-exposed environment to one that interacts extensively with helix 12. Hydrogen-deuterium exchange mass spectrometry reveals that the dynamics of helices 3, 11, and 12 are significantly decreased when the two rexinoids are bound to the receptor. When the pan-agonist 9-cis-retinoic acid is bound to the receptor, only the dynamics of helices 3 and 11 are reduced. The four structural changes are conserved in all x-ray structures of the RXR ligand-binding domain in the presence of agonist and coactivator peptide. They serve as hallmarks for how RXR changes conformation and dynamics in the presence of agonist and coactivator to initiate signaling.
- From the Departments of Biochemistry and Molecular Genetics.
Organizational Affiliation: