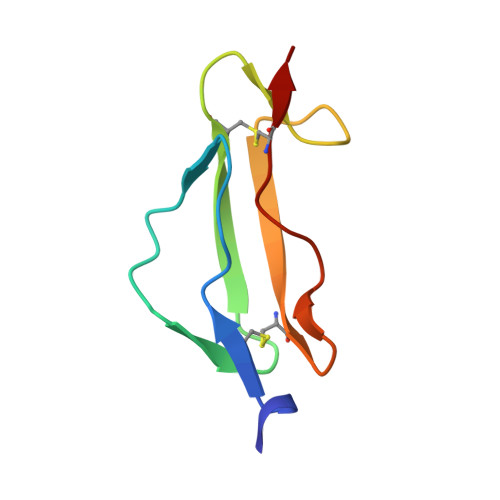
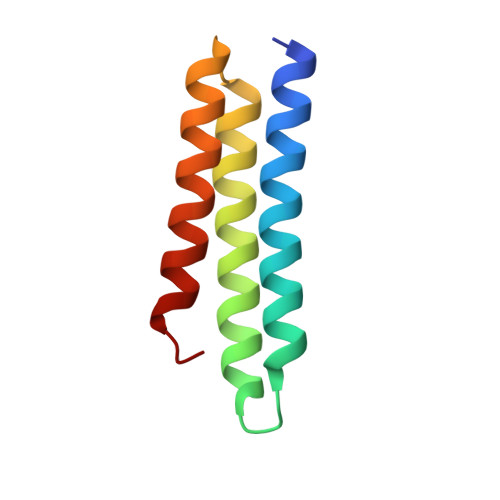
Structural determinants of host specificity of complement Factor H recruitment by Streptococcus pneumoniae.
Achila, D., Liu, A., Banerjee, R., Li, Y., Martinez-Hackert, E., Zhang, J.R., Yan, H.(2015) Biochem J 465: 325-335
- PubMed: 25330773
- DOI: https://doi.org/10.1042/BJ20141069
- Primary Citation of Related Structures:
2M6U, 4K12 - PubMed Abstract:
Many human pathogens have strict host specificity, which affects not only their epidemiology but also the development of animal models and vaccines. Complement Factor H (FH) is recruited to pneumococcal cell surface in a human-specific manner via the N-terminal domain of the pneumococcal protein virulence factor choline-binding protein A (CbpAN). FH recruitment enables Streptococcus pneumoniae to evade surveillance by human complement system and contributes to pneumococcal host specificity. The molecular determinants of host specificity of complement evasion are unknown. In the present study, we show that a single human FH (hFH) domain is sufficient for tight binding of CbpAN, present the crystal structure of the complex and identify the critical structural determinants for host-specific FH recruitment. The results offer new approaches to the development of better animal models for pneumococcal infection and redesign of the virulence factor for pneumococcal vaccine development and reveal how FH recruitment can serve as a mechanism for both pneumococcal complement evasion and adherence.
- *Department of Biochemistry and Molecular Biology, Michigan State University, 603 Wilson Road, East Lansing, MI 48824, U.S.A.
Organizational Affiliation: