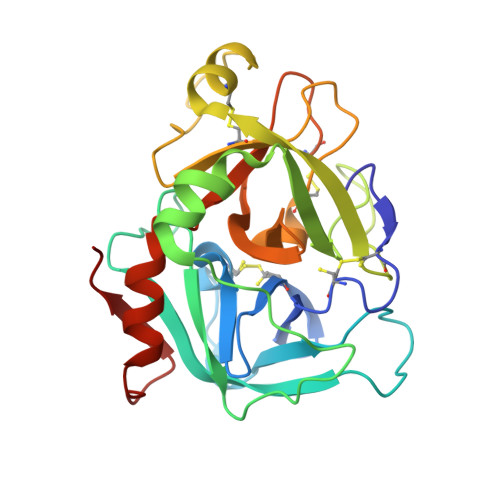
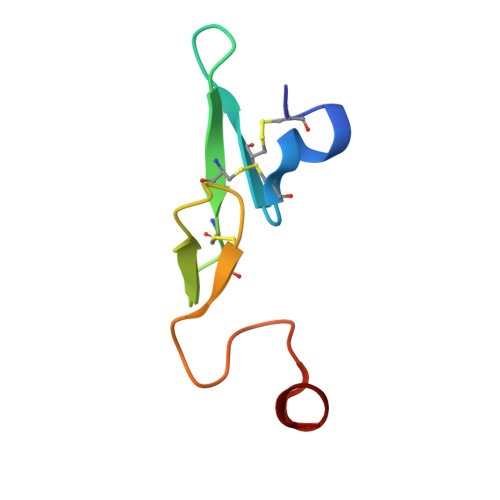
Nonbenzamidine acylsulfonamide tissue factor-factor VIIa inhibitors.
Glunz, P.W., Zhang, X., Zou, Y., Delucca, I., Nirschl, A.H., Cheng, X., Weigelt, C.A., Cheney, D.L., Wei, A., Anumula, R., Luettgen, J.M., Rendina, A.R., Harpel, M., Luo, G., Knabb, R., Wong, P.C., Wexler, R.R., Priestley, E.S.(2013) Bioorg Med Chem Lett 23: 5244-5248
- PubMed: 23845220
- DOI: https://doi.org/10.1016/j.bmcl.2013.06.027
- Primary Citation of Related Structures:
4JYU, 4JYV - PubMed Abstract:
Aminoisoquinoline and isoquinoline groups have successfully replaced the more basic P1 benzamidine group of an acylsulfonamide factor VIIa inhibitor. Inhibitory activity was optimized by the identification of additional hydrophobic and hydrophilic P' binding interactions. The molecular details of these interactions were elucidated by X-ray crystallography and molecular modeling. We also show that decreasing the basicity of the P1 group results in improved oral bioavailability in this chemotype.
- Bristol-Myers Squibb R&D, Pennington, NJ 08534, United States. peter.glunz@bms.com
Organizational Affiliation: