Distinguishing drug binding pockets on proteins by complementary biophysical and biological methods
Laplante, S.R., Coulombe, R.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Genome polyprotein | 576 | Hepatitis C virus isolate HC-J4 | Mutation(s): 0 Gene Names: NS5B EC: 3.4.22 (PDB Primary Data), 3.4.21.98 (PDB Primary Data), 3.6.1.15 (PDB Primary Data), 3.6.4.13 (PDB Primary Data), 2.7.7.48 (PDB Primary Data) | 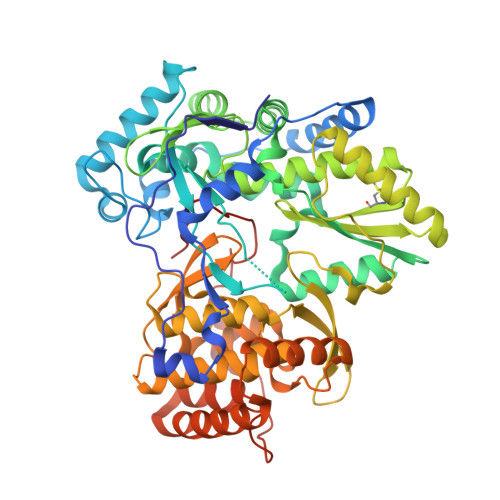 | |
UniProt | |||||
Find proteins for O92972 (Hepatitis C virus genotype 1b (strain HC-J4)) Explore O92972 Go to UniProtKB: O92972 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O92972 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NN3 Query on NN3 | C [auth A] | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID C22 H27 N O3 S RZXQBIKGWSLVEK-JCNLHEQBSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 106.26 | α = 90 |
| b = 108.03 | β = 90 |
| c = 135.13 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| CNX | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| CNX | phasing |