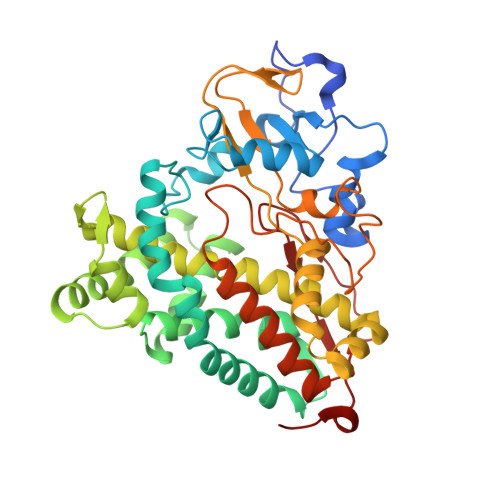
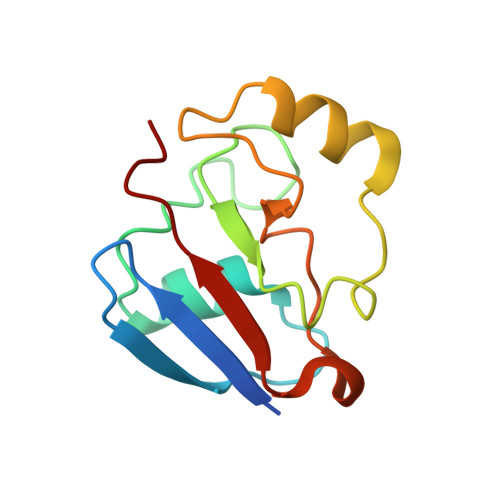
Structural basis for effector control and redox partner recognition in cytochrome P450.
Tripathi, S., Li, H., Poulos, T.L.(2013) Science 340: 1227-1230
- PubMed: 23744947
- DOI: https://doi.org/10.1126/science.1235797
- Primary Citation of Related Structures:
4JWS, 4JWU, 4JX1 - PubMed Abstract:
Cytochromes P450 catalyze a variety of monooxygenase reactions that require electron transfer from redox partners. Although the structure of many P450s and a small handful of redox partners are known, there is very little structural information available on redox complexes, thus leaving a gap in our understanding on the control of P450-redox partner interactions. We have solved the crystal structure of oxidized and reduced P450cam complexed with its redox partner, putidaredoxin (Pdx), to 2.2 and 2.09 angstroms, respectively. It was anticipated that Pdx would favor closed substrate-bound P450cam, which differs substantially from the open conformer, but instead we found that Pdx favors the open state. These new structures indicate that the effector role of Pdx is to shift P450cam toward the open conformation, which enables the establishment of a water-mediated H-bonded network, which is required for proton-coupled electron transfer.
- Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA 92697-3900, USA.
Organizational Affiliation: