A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms.
Metzger, M.B., Liang, Y.H., Das, R., Mariano, J., Li, S., Li, J., Kostova, Z., Byrd, R.A., Ji, X., Weissman, A.M.(2013) Mol Cell 50: 516-527
- PubMed: 23665230
- DOI: https://doi.org/10.1016/j.molcel.2013.04.004
- Primary Citation of Related Structures:
4JQU - PubMed Abstract:
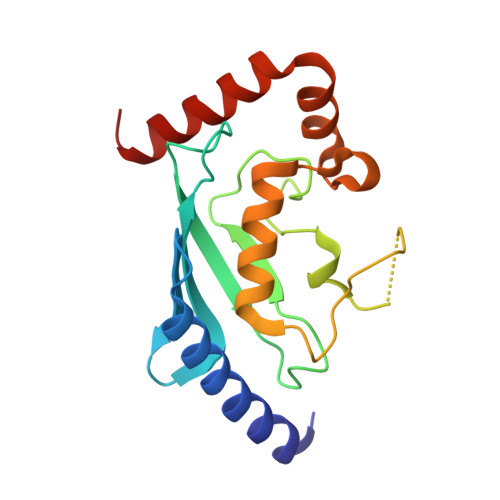
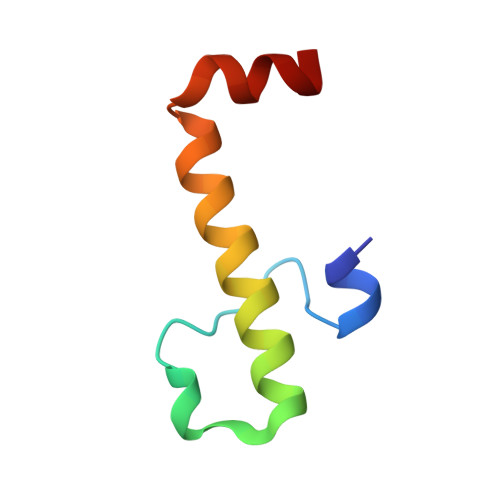
Cue1p is an integral component of yeast endoplasmic reticulum (ER)-associated degradation (ERAD) ubiquitin ligase (E3) complexes. It tethers the ERAD ubiquitin-conjugating enzyme (E2), Ubc7p, to the ER and prevents its degradation, and also activates Ubc7p via unknown mechanisms. We have now determined the crystal structure of the Ubc7p-binding region (U7BR) of Cue1p with Ubc7p. The U7BR is a unique E2-binding domain that includes three α-helices that interact extensively with the "backside" of Ubc7p. Residues essential for E2 binding are also required for activation of Ubc7p and for ERAD. We establish that the U7BR stimulates both RING-independent and RING-dependent ubiquitin transfer from Ubc7p. Moreover, the U7BR enhances ubiquitin-activating enzyme (E1)-mediated charging of Ubc7p with ubiquitin. This demonstrates that an essential component of E3 complexes can simultaneously bind to E2 and enhance its loading with ubiquitin. These findings provide mechanistic insights into how ubiquitination can be stimulated.
- Laboratory of Protein Dynamics and Signaling, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702-1201, USA.
Organizational Affiliation: