Differential regulation by ppGpp versus pppGpp in Escherichia coli.
Mechold, U., Potrykus, K., Murphy, H., Murakami, K.S., Cashel, M.(2013) Nucleic Acids Res 41: 6175-6189
- PubMed: 23620295
- DOI: https://doi.org/10.1093/nar/gkt302
- Primary Citation of Related Structures:
4JK1, 4JK2 - PubMed Abstract:
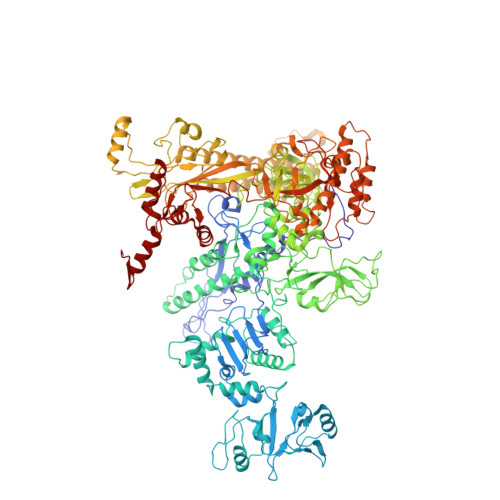
Both ppGpp and pppGpp are thought to function collectively as second messengers for many complex cellular responses to nutritional stress throughout biology. There are few indications that their regulatory effects might be different; however, this question has been largely unexplored for lack of an ability to experimentally manipulate the relative abundance of ppGpp and pppGpp. Here, we achieve preferential accumulation of either ppGpp or pppGpp with Escherichia coli strains through induction of different Streptococcal (p)ppGpp synthetase fragments. In addition, expression of E. coli GppA, a pppGpp 5'-gamma phosphate hydrolase that converts pppGpp to ppGpp, is manipulated to fine tune differential accumulation of ppGpp and pppGpp. In vivo and in vitro experiments show that pppGpp is less potent than ppGpp with respect to regulation of growth rate, RNA/DNA ratios, ribosomal RNA P1 promoter transcription inhibition, threonine operon promoter activation and RpoS induction. To provide further insights into regulation by (p)ppGpp, we have also determined crystal structures of E. coli RNA polymerase-σ(70) holoenzyme with ppGpp and pppGpp. We find that both nucleotides bind to a site at the interface between β' and ω subunits.
- Laboratory of Molecular Genetics, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: