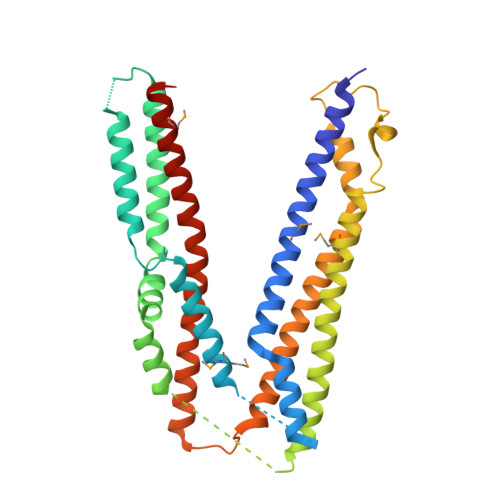
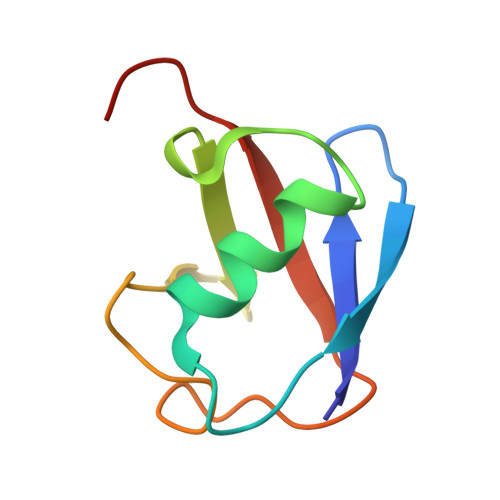
The yeast alix homolog bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes.
Pashkova, N., Gakhar, L., Winistorfer, S.C., Sunshine, A.B., Rich, M., Dunham, M.J., Yu, L., Piper, R.C.(2013) Dev Cell 25: 520-533
- PubMed: 23726974
- DOI: https://doi.org/10.1016/j.devcel.2013.04.007
- Primary Citation of Related Structures:
4JIO, 4JJY - PubMed Abstract:
Sorting of ubiquitinated membrane proteins into lumenal vesicles of multivesicular bodies is mediated by the Endosomal Sorting Complex Required for Transport (ESCRT) apparatus and accessory proteins such as Bro1, which recruits the deubiquitinating enzyme Doa4 to remove ubiquitin from cargo. Here we propose that Bro1 works as a receptor for the selective sorting of ubiquitinated cargoes. We found synthetic genetic interactions between BRO1 and ESCRT-0, suggesting that Bro1 functions similarly to ESCRT-0. Multiple structural approaches demonstrated that Bro1 binds ubiquitin via the N-terminal trihelical arm of its middle V domain. Mutants of Bro1 that lack the ability to bind Ub were dramatically impaired in their ability to sort Ub-cargo membrane proteins, but only when combined with hypomorphic alleles of ESCRT-0. These data suggest that Bro1 and other Bro1 family members function in parallel with ESCRT-0 to recognize and sort Ub-cargoes.
- Department of Molecular Physiology and Biophysics, University of Iowa, Iowa City, IA 52242, USA.
Organizational Affiliation: