Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Pseudovibrio sp.
Hegde, R.P., Toro, R., Burley, S.K., Almo, S.C., Ramagopal, U.A., New York Structural Genomics Research Consortium (NYSGRC)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | 384 | Pseudovibrio sp. JE062 | Mutation(s): 0 Gene Names: PJE062_89, PJE062_892 | 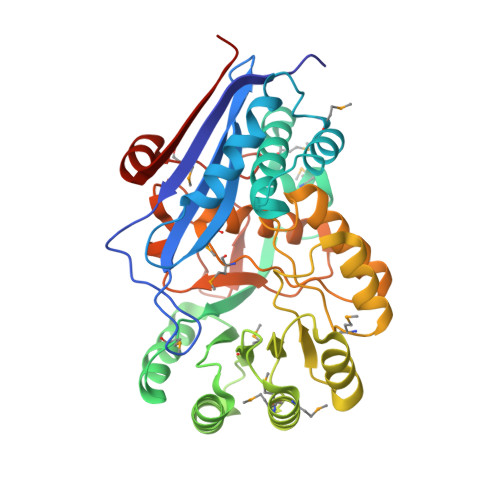 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B, C, D, E A, B, C, D, E, F, G, H | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 201.793 | α = 90 |
| b = 117.313 | β = 99.77 |
| c = 143.127 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALA | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| CBASS | data collection |
| MOSFLM | data reduction |
| MOLREP | phasing |