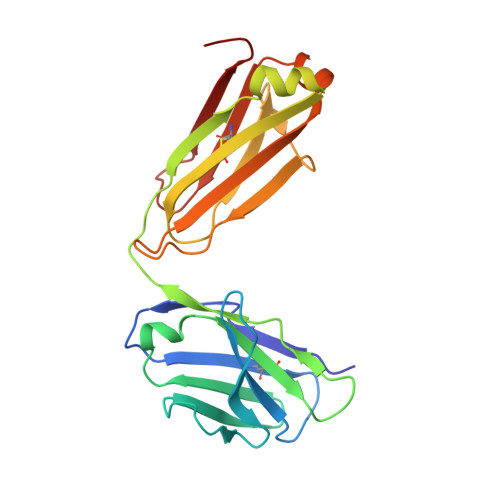
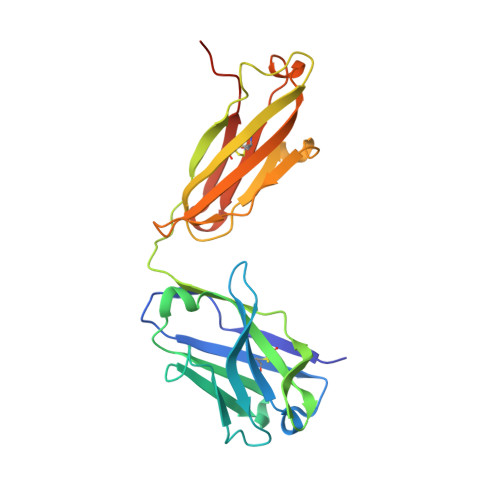
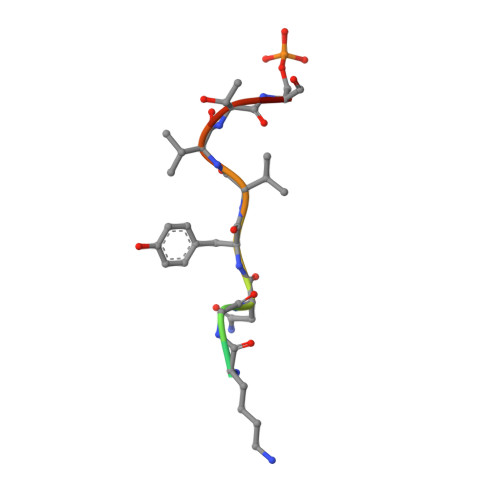
Nature-inspired design of motif-specific antibody scaffolds.
Koerber, J.T., Thomsen, N.D., Hannigan, B.T., Degrado, W.F., Wells, J.A.(2013) Nat Biotechnol 31: 916-921
- PubMed: 23955275
- DOI: https://doi.org/10.1038/nbt.2672
- Primary Citation of Related Structures:
4JFX, 4JFY, 4JFZ, 4JG0, 4JG1 - PubMed Abstract:
Aberrant changes in post-translational modifications (PTMs) such as phosphate groups underlie a majority of human diseases. However, detection and quantification of PTMs for diagnostic or biomarker applications often require PTM-specific monoclonal antibodies (mAbs), which are challenging to generate using traditional antibody-selection methods. Here we outline a general strategy for producing synthetic, PTM-specific mAbs by engineering a motif-specific 'hot spot' into an antibody scaffold. Inspired by a natural phosphate-binding motif, we designed and selected mAb scaffolds with hot spots specific for phosphoserine, phosphothreonine or phosphotyrosine. Crystal structures of the phospho-specific mAbs revealed two distinct modes of phosphoresidue recognition. Our data suggest that each hot spot functions independently of the surrounding scaffold, as phage display antibody libraries using these scaffolds yielded >50 phospho- and target-specific mAbs against 70% of target peptides. Our motif-specific scaffold strategy may provide a general solution for rapid, robust development of anti-PTM mAbs for signaling, diagnostic and therapeutic applications.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, California, USA.
Organizational Affiliation: