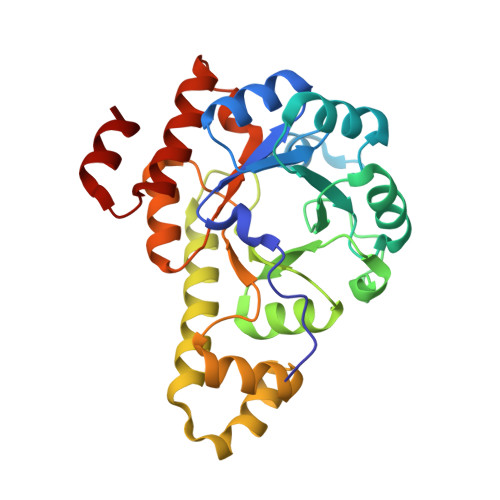
Crystal structure of Mycobacterium tuberculosis Rv2606c: a pyridoxal biosynthesis lyase.
Kim, S., Kim, K.J.(2013) Biochem Biophys Res Commun 435: 255-259
- PubMed: 23643787
- DOI: https://doi.org/10.1016/j.bbrc.2013.04.068
- Primary Citation of Related Structures:
4JDY - PubMed Abstract:
Tuberculosis is a lethal infectious disease caused by Mycobacterium tuberculosis. We determined the crystal structure of Rv2606c, a potential pyridoxal biosynthesis lyase (PdxS), from M. tuberculosis H37Rv at 1.8 Å resolution. The overall structure of the protein, composed of a (β/α)8-barrel and two small 310-helices, was quite similar to those of other PdxS proteins. A glycerol molecule was observed to be bound at the active site of the Rv2606c structure through interactions with the conserved residues of Asp29 and Lys86, providing information regarding the potential active site and the substrate-binding environment of the protein. The interface for Rv2606c dodecamerization, which is primarily mediated by salt bridges and hydrophobic interactions, was quite different from those of other PdxS proteins. Furthermore, we observed that the Rv2606c and Rv2604c form a stable complex, suggesting that these proteins might function as PdxS and PdxT in M. tuberculosis.
- School of Life Science and Biotechnology, Kyungpook National University, Republic of Korea.
Organizational Affiliation: