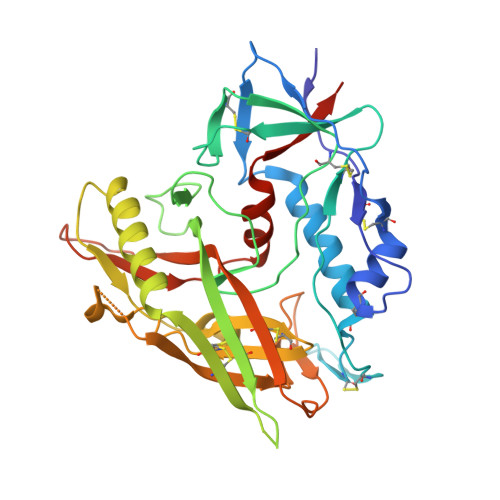
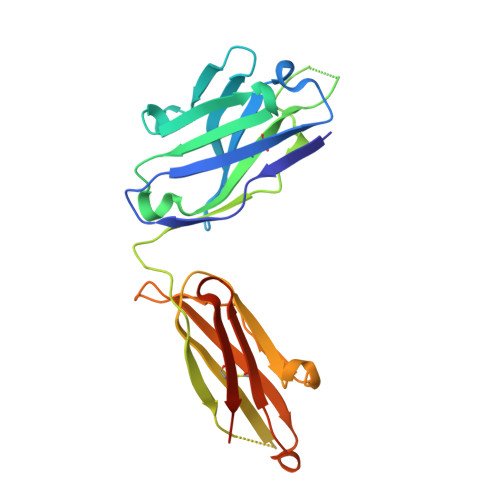
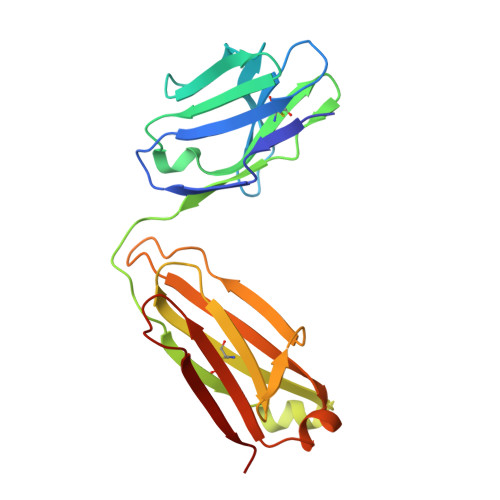
Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Scharf, L., West, A.P., Gao, H., Lee, T., Scheid, J.F., Nussenzweig, M.C., Bjorkman, P.J., Diskin, R.(2013) Proc Natl Acad Sci U S A 110: 6049-6054
- PubMed: 23524883
- DOI: https://doi.org/10.1073/pnas.1303682110
- Primary Citation of Related Structures:
4JDT, 4JDV - PubMed Abstract:
Efforts to design an effective antibody-based vaccine against HIV-1 would benefit from understanding how germ-line B-cell receptors (BCRs) recognize the HIV-1 gp120/gp41 envelope spike. Potent VRC01-like (PVL) HIV-1 antibodies derived from the VH1-2*02 germ-line allele target the conserved CD4 binding site on gp120. A bottleneck for design of immunogens capable of eliciting PVL antibodies is that VH1-2*02 germ-line BCR interactions with gp120 are uncharacterized. Here, we report the structure of a VH1-2*02 germ-line antibody alone and a germ-line heavy-chain/mature light-chain chimeric antibody complexed with HIV-1 gp120. VH1-2*02 residues make extensive contacts with the gp120 outer domain, including all PVL signature and CD4 mimicry interactions, but not critical CDRH3 contacts with the gp120 inner domain and bridging sheet that are responsible for the improved potency of NIH45-46 over closely related clonal variants, such as VRC01. Our results provide insight into initial recognition of HIV-1 by VH1-2*02 germ-line BCRs and may facilitate the design of immunogens tailored to engage and stimulate broad and potent CD4 binding site antibodies.
- Division of Biology, California Institute of Technology, Pasadena, CA 91125, USA.
Organizational Affiliation: