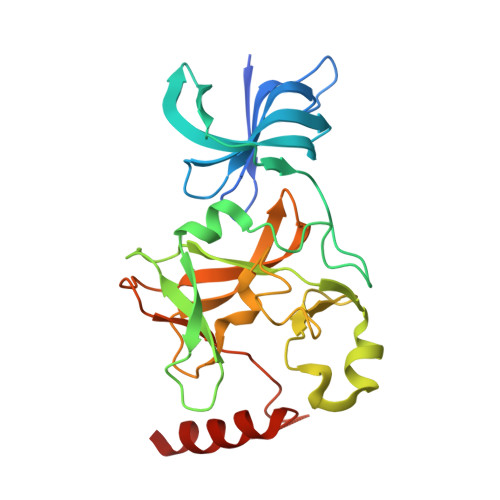
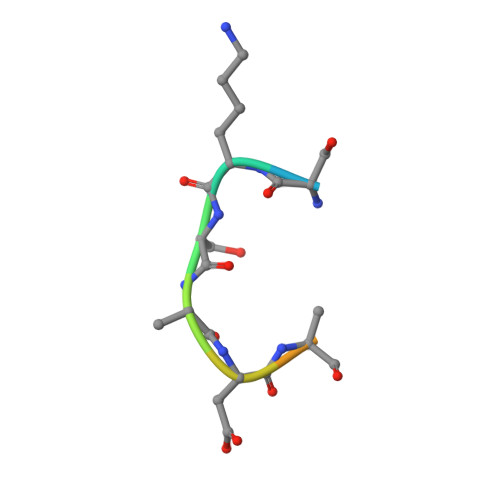
Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
Horowitz, S., Dirk, L.M., Yesselman, J.D., Nimtz, J.S., Adhikari, U., Mehl, R.A., Scheiner, S., Houtz, R.L., Al-Hashimi, H.M., Trievel, R.C.(2013) J Am Chem Soc 135: 15536-15548
- PubMed: 24093804
- DOI: https://doi.org/10.1021/ja407140k
- Primary Citation of Related Structures:
4J7I, 4J83, 4J8O - PubMed Abstract:
S-adenosylmethionine (AdoMet)-based methylation is integral to metabolism and signaling. AdoMet-dependent methyltransferases belong to multiple distinct classes and share a catalytic mechanism that arose through convergent evolution; however, fundamental determinants underlying this shared methyl transfer mechanism remain undefined. A survey of high-resolution crystal structures reveals that unconventional carbon-oxygen (CH···O) hydrogen bonds coordinate the AdoMet methyl group in different methyltransferases irrespective of their class, active site structure, or cofactor binding conformation. Corroborating these observations, quantum chemistry calculations demonstrate that these charged interactions formed by the AdoMet sulfonium cation are stronger than typical CH···O hydrogen bonds. Biochemical and structural studies using a model lysine methyltransferase and an active site mutant that abolishes CH···O hydrogen bonding to AdoMet illustrate that these interactions are important for high-affinity AdoMet binding and transition-state stabilization. Further, crystallographic and NMR dynamics experiments of the wild-type enzyme demonstrate that the CH···O hydrogen bonds constrain the motion of the AdoMet methyl group, potentially facilitating its alignment during catalysis. Collectively, the experimental findings with the model methyltransferase and structural survey imply that methyl CH···O hydrogen bonding represents a convergent evolutionary feature of AdoMet-dependent methyltransferases, mediating a universal mechanism for methyl transfer.
- Howard Hughes Medical Institute , Ann Arbor, Michigan 48109, United States.
Organizational Affiliation: