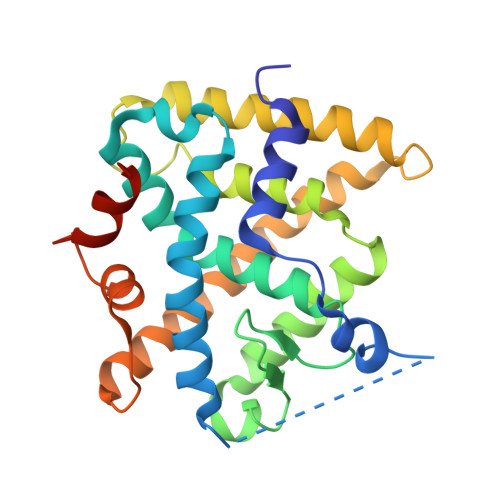
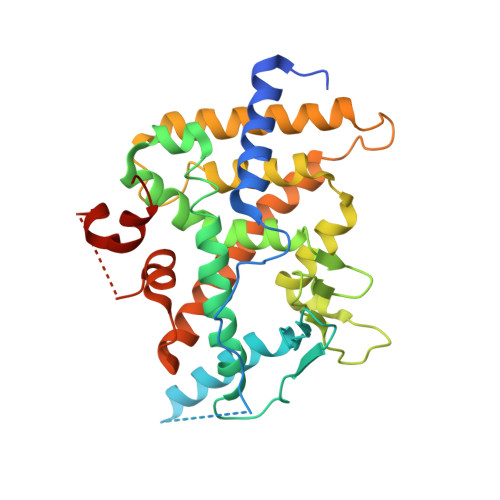
Structural and Functional Analysis of the Human Nuclear Xenobiotic Receptor PXR in Complex with RXRalpha.
Wallace, B.D., Betts, L., Talmage, G., Pollet, R.M., Holman, N.S., Redinbo, M.R.(2013) J Mol Biology 425: 2561-2577
- PubMed: 23602807
- DOI: https://doi.org/10.1016/j.jmb.2013.04.012
- Primary Citation of Related Structures:
4J5W, 4J5X - PubMed Abstract:
The human nuclear xenobiotic receptor PXR recognizes a range of potentially harmful drugs and endobiotic chemicals but must complex with the nuclear receptor RXRα to control the expression of numerous drug metabolism genes. To date, the structural basis and functional consequences of this interaction have remained unclear. Here we present 2.8-Å-resolution crystal structures of the heterodimeric complex formed between the ligand-binding domains of human PXR and RXRα. These structures establish that PXR and RXRα form a heterotetramer unprecedented in the nuclear receptor family of ligand-regulated transcription factors. We further show that both PXR and RXRα bind to the transcriptional coregulator SRC-1 with higher affinity when they are part of the PXR/RXRα heterotetramer complex than they do when each ligand-binding domain is examined alone. Furthermore, we purify the full-length forms of each receptor from recombinant bacterial expression systems and characterize their interactions with a range of direct and everted repeat DNA elements. Taken together, these data advance our understanding of PXR, the master regulator of drug metabolism gene expression in humans, in its functional partnership with RXRα.
- Department of Chemistry, Biochemistry and Microbiology, University of North Carolina at Chapel Hill, 250 Bell Tower Drive, Chapel Hill, NC 27599, USA.
Organizational Affiliation: