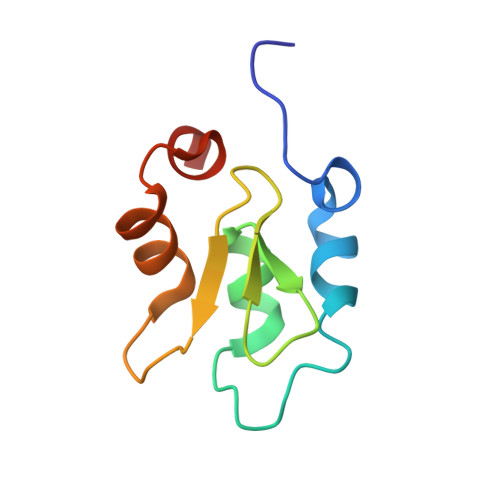
The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Lukacs, C., Belunis, C., Crowther, R., Danho, W., Gao, L., Goggin, B., Janson, C.A., Li, S., Remiszewski, S., Schutt, A., Thakur, M.K., Singh, S.K., Swaminathan, S., Pandey, R., Tyagi, R., Gosu, R., Kamath, A.V., Kuglstatter, A.(2013) Acta Crystallogr D Biol Crystallogr 69: 1717-1725
- PubMed: 23999295
- DOI: https://doi.org/10.1107/S0907444913016284
- Primary Citation of Related Structures:
4J3Y, 4J44, 4J45, 4J46, 4J47, 4J48 - PubMed Abstract:
XIAP, a member of the inhibitor of apoptosis family of proteins, is a critical regulator of apoptosis. Inhibition of the BIR domain-caspase interaction is a promising approach towards treating cancer. Previous work has been directed towards inhibiting the BIR3-caspase-9 interaction, which blocks the intrinsic apoptotic pathway; selectively inhibiting the BIR2-caspase-3 interaction would also block the extrinsic pathway. The BIR2 domain of XIAP has successfully been crystallized; peptides and small-molecule inhibitors can be soaked into these crystals, which diffract to high resolution. Here, the BIR2 apo crystal structure and the structures of five BIR2-tetrapeptide complexes are described. The structural flexibility observed on comparing these structures, along with a comparison with XIAP BIR3, affords an understanding of the structural elements that drive selectivity between BIR2 and BIR3 and which can be used to design BIR2-selective inhibitors.
- Discovery Technologies, Hoffmann-La Roche, 340 Kingsland Street, Nutley, NJ 07110, USA.
Organizational Affiliation: