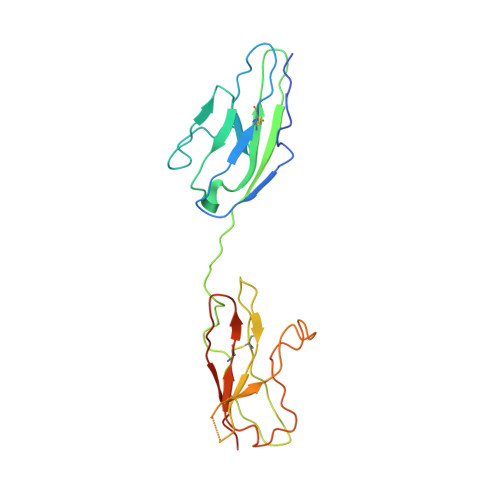
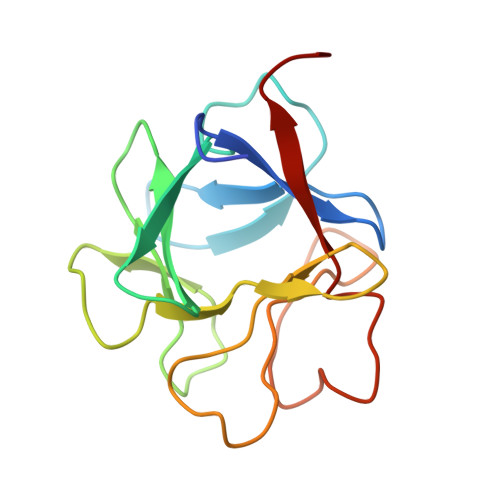
Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling.
Herbert, C., Schieborr, U., Saxena, K., Juraszek, J., De Smet, F., Alcouffe, C., Bianciotto, M., Saladino, G., Sibrac, D., Kudlinzki, D., Sreeramulu, S., Brown, A., Rigon, P., Herault, J.P., Lassalle, G., Blundell, T.L., Rousseau, F., Gils, A., Schymkowitz, J., Tompa, P., Herbert, J.M., Carmeliet, P., Gervasio, F.L., Schwalbe, H., Bono, F.(2013) Cancer Cell 23: 489-501
- PubMed: 23597563
- DOI: https://doi.org/10.1016/j.ccr.2013.02.018
- Primary Citation of Related Structures:
4J23 - PubMed Abstract:
The fibroblast growth factor (FGF)/fibroblast growth factor receptor (FGFR) signaling network plays an important role in cell growth, survival, differentiation, and angiogenesis. Deregulation of FGFR signaling can lead to cancer development. Here, we report an FGFR inhibitor, SSR128129E (SSR), that binds to the extracellular part of the receptor. SSR does not compete with FGF for binding to FGFR but inhibits FGF-induced signaling linked to FGFR internalization in an allosteric manner, as shown by crystallography studies, nuclear magnetic resonance, Fourier transform infrared spectroscopy, molecular dynamics simulations, free energy calculations, structure-activity relationship analysis, and FGFR mutagenesis. Overall, SSR is a small molecule allosteric inhibitor of FGF/FGFR signaling, acting via binding to the extracellular part of the FGFR.
- E2C and LGCR-SDI Department, Sanofi Research and Development, 31100 Toulouse, France.
Organizational Affiliation: