Seed Sequence-Matched Controls Reveal Limitations of Small Interfering RNA Knockdown in Functional and Structural Studies of Hepatitis C Virus NS5A-MOBKL1B Interaction.
Chung, H.Y., Gu, M., Buehler, E., MacDonald, M.R., Rice, C.M.(2014) J Virol 88: 11022-11033
- PubMed: 25031347
- DOI: https://doi.org/10.1128/JVI.01582-14
- Primary Citation of Related Structures:
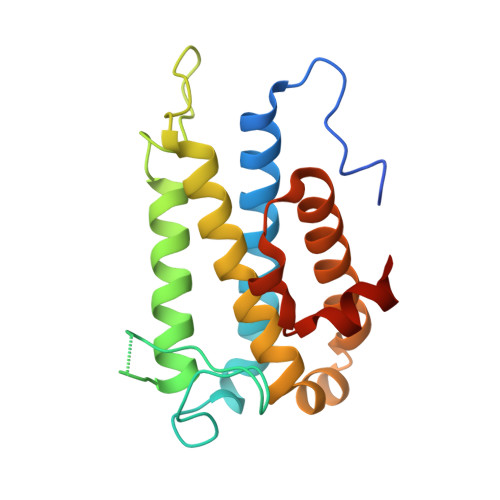
4J1V - PubMed Abstract:
Hepatitis C virus (HCV) is a widespread human pathogen causing liver cirrhosis and cancer. Similar to the case for other viruses, HCV depends on host and viral factors to complete its life cycle. We used proteomic and yeast two-hybrid approaches to elucidate host factors involved in HCV nonstructural protein NS5A function and found that MOBKL1B interacts with NS5A. Initial experiments with small interfering RNA (siRNA) knockdown suggesting a role in HCV replication led us to examine the interaction using biochemical and structural approaches. As revealed by a cocrystal structure of a core MOBKL1B-NS5A peptide complex at 1.95 Å, NS5A binds to a hydrophobic patch on the MOBKL1B surface. Biosensor binding assays identified a highly conserved, 18-amino-acid binding site in domain II of NS5A, which encompasses residues implicated in cyclophilin A (CypA)-dependent HCV RNA replication. However, a CypA-independent HCV variant had reduced replication in MOBKL1B knockdown cells, even though its NS5A does not interact with MOBKL1B. These discordant results prompted more extensive studies of MOBKL1B gene knockdowns, which included additional siRNAs and specifically matched seed sequence siRNA controls. We found that reduced virus replication after treating cells with MOBKL1B siRNA was actually due to off-target inhibition, which indicated that the initial finding of virus replication dependence on the MOBKL1B-NS5A interaction was incorrect. Ultimately, using several approaches, we found no relationship of the MOBKL1B-NS5A interaction to virus replication. These findings collectively serve as a reminder to investigators and scientific reviewers of the pervasive impact of siRNA off-target effects on interpretation of biological data. Our study illustrates an underappreciated shortcoming of siRNA gene knockdown technology. We initially identified a cellular protein, MOBKL1B, as a binding partner with the NS5A protein of hepatitis C virus (HCV). MOBKL1B siRNA, but not irrelevant RNA, treatment was associated with both reduced virus replication and the absence of MOBKL1B. Believing that HCV replication depended on the MOBKL1B-NS5A interaction, we carried out structural and biochemical analyses. Unexpectedly, an HCV variant lacking the MOBKL1B-NS5A interaction could not replicate after cells were treated with MOBKL1B siRNA. By repeating the MOBKL1B siRNA knockdowns and including seed sequence-matched siRNA instead of irrelevant siRNA as a control, we found that the MOBKL1B siRNAs utilized had off-target inhibitory effects on virus replication. Collectively, our results suggest that stricter controls must be utilized in all RNA interference (RNAi)-mediated gene knockdown experiments to ensure sound conclusions and a reliable scientific knowledge database.
- Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, New York, USA.
Organizational Affiliation: