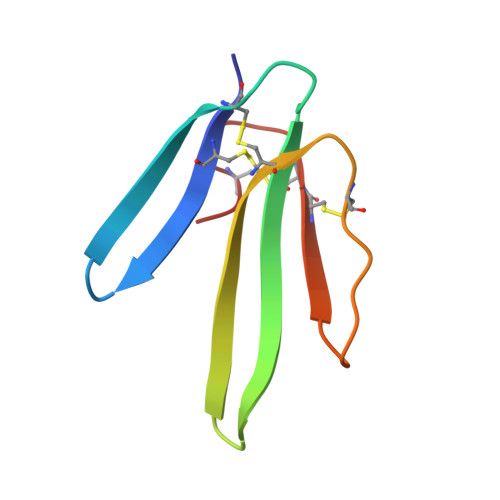
Crystallization of recombinant green mamba rho-Da1a toxin during a lyophilization procedure and its structure determination.
Maiga, A., Vera, L., Marchetti, C., Lorphelin, A., Bellanger, L., Mourier, G., Servent, D., Gilles, N., Stura, E.A.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 704-709
- PubMed: 23722859
- DOI: https://doi.org/10.1107/S1744309113011470
- Primary Citation of Related Structures:
4IYE - PubMed Abstract:
ρ-Da1a toxin from eastern green mamba (Dendroaspis angusticeps) venom is a polypeptide of 65 amino acids with a strong affinity for the G-protein-coupled α(1A)-adrenoceptor. This neurotoxin has been crystallized from resolubilized lyophilized powder, but the best crystals grew spontaneously during lyophilization. The crystals belonged to the trigonal space group P3(1)21, with unit-cell parameters a = b = 37.37, c = 66.05 Å, and diffracted to 1.95 Å resolution. The structure solved by molecular replacement showed strong similarities to green mamba muscarinic toxins.
- CEA, DSV, iBiTec-S, Service d'Ingénierie Moléculaire des Protéines, 91191 Gif-sur-Yvette, France.
Organizational Affiliation: