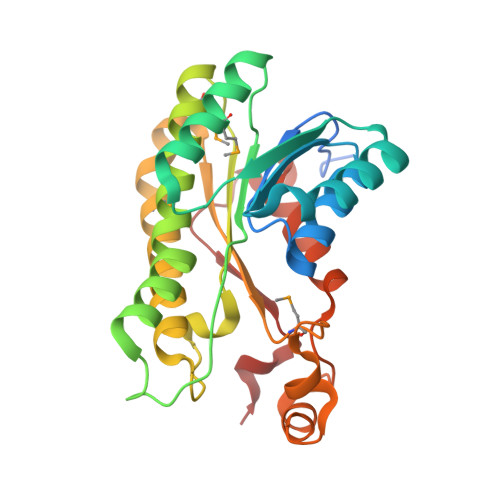
Structure of a short-chain dehydrogenase/reductase (SDR) within a genomic island from a clinical strain of Acinetobacter baumannii.
Shah, B.S., Tetu, S.G., Harrop, S.J., Paulsen, I.T., Mabbutt, B.C.(2014) Acta Crystallogr F Struct Biol Commun 70: 1318-1323
- PubMed: 25286932
- DOI: https://doi.org/10.1107/S2053230X14019785
- Primary Citation of Related Structures:
4IUY - PubMed Abstract:
Over 15% of the genome of an Australian clinical isolate of Acinetobacter baumannii occurs within genomic islands. An uncharacterized protein encoded within one island feature common to this and other International Clone II strains has been studied by X-ray crystallography. The 2.4 Å resolution structure of SDR-WM99c reveals it to be a new member of the classical short-chain dehydrogenase/reductase (SDR) superfamily. The enzyme contains a nucleotide-binding domain and, like many other SDRs, is tetrameric in form. The active site contains a catalytic tetrad (Asn117, Ser146, Tyr159 and Lys163) and water molecules occupying the presumed NADP cofactor-binding pocket. An adjacent cleft is capped by a relatively mobile helical subdomain, which is well positioned to control substrate access.
- Department of Chemistry and Biomolecular Sciences, Macquarie University, Research Park Drive, Sydney, NSW 2109, Australia.
Organizational Affiliation: