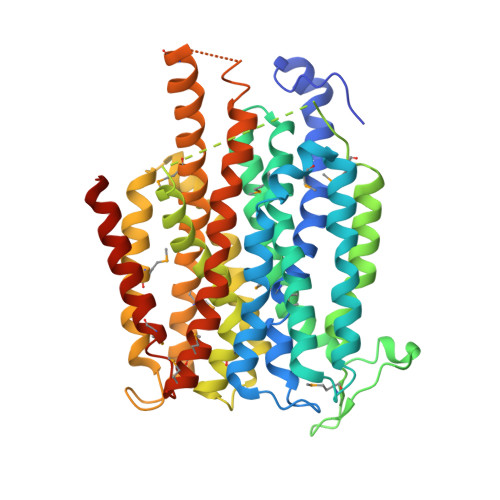
Structure and mechanism of a nitrate transporter.
Yan, H., Huang, W., Yan, C., Gong, X., Jiang, S., Zhao, Y., Wang, J., Shi, Y.(2013) Cell Rep 3: 716-723
- PubMed: 23523348
- DOI: https://doi.org/10.1016/j.celrep.2013.03.007
- Primary Citation of Related Structures:
4IU8, 4IU9 - PubMed Abstract:
The nitrate/nitrite transporters NarK and NarU play an important role in nitrogen homeostasis in bacteria and belong to the nitrate/nitrite porter family (NNP) of the major facilitator superfamily (MFS) fold. The structure and functional mechanism of NarK and NarU remain unknown. Here, we report the crystal structure of NarU at a resolution of 3.1 Å and systematic biochemical characterization. The two molecules of NarU in an asymmetric unit exhibit two distinct conformational states: occluded and partially inward-open. The substrate molecule nitrate appears to be coordinated by four highly conserved, charged, or polar amino acids. Structural and biochemical analyses allowed the identification of key amino acids that are involved in substrate gating and transport. The observed conformational differences of NarU, together with unique sequence features of the NNP family transporters, suggest a transport mechanism that might deviate from the canonical rocker-switch model.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: