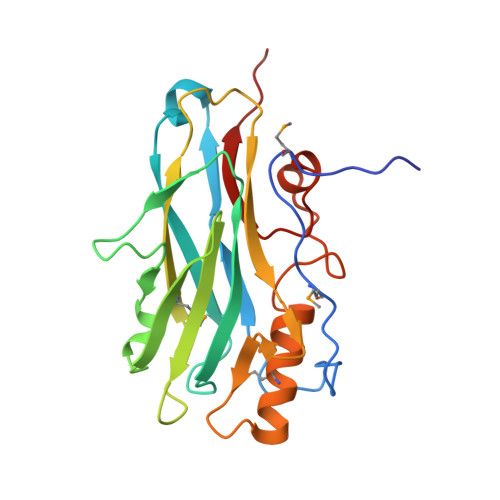
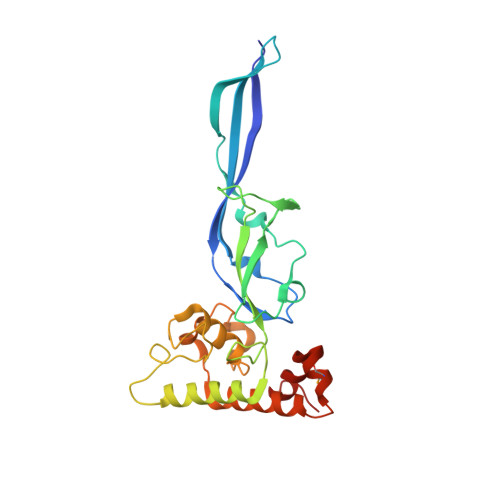
Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
Salama-Alber, O., Jobby, M.K., Chitayat, S., Smith, S.P., White, B.A., Shimon, L.J., Lamed, R., Frolow, F., Bayer, E.A.(2013) J Biological Chem 288: 16827-16838
- PubMed: 23580648
- DOI: https://doi.org/10.1074/jbc.M113.466672
- Primary Citation of Related Structures:
4IU2, 4IU3 - PubMed Abstract:
The rumen bacterium Ruminococcus flavefaciens produces a highly organized multienzyme cellulosome complex that plays a key role in the degradation of plant cell wall polysaccharides, notably cellulose. The R. flavefaciens cellulosomal system is anchored to the bacterial cell wall through a relatively small ScaE scaffoldin subunit, which bears a single type IIIe cohesin responsible for the attachment of two major dockerin-containing scaffoldin proteins, ScaB and the cellulose-binding protein CttA. Although ScaB recruits the catalytic machinery onto the complex, CttA mediates attachment of the bacterial substrate via its two putative carbohydrate-binding modules. In an effort to understand the structural basis for assembly and cell surface attachment of the cellulosome in R. flavefaciens, we determined the crystal structure of the high affinity complex (Kd = 20.83 nM) between the cohesin module of ScaE (CohE) and its cognate X-dockerin (XDoc) modular dyad from CttA at 1.97-Å resolution. The structure reveals an atypical calcium-binding loop containing a 13-residue insert. The results further pinpoint two charged specificity-related residues on the surface of the cohesin module that are responsible for specific versus promiscuous cross-strain binding of the dockerin module. In addition, a combined functional role for the three enigmatic dockerin inserts was established whereby these extraneous segments serve as structural buttresses that reinforce the stalklike conformation of the X-module, thus segregating its tethered complement of cellulosomal components from the cell surface. The novel structure of the RfCohE-XDoc complex sheds light on divergent dockerin structure and function and provides insight into the specificity features of the type IIIe cohesin-dockerin interaction.
- Departments of Biological Chemistry, Rehovot 76100, Israel.
Organizational Affiliation: