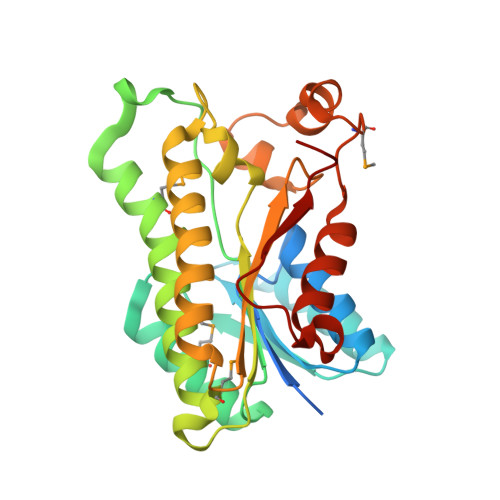
Crystal structure of oxidoreductase from Polaromonas sp. in NADP bound form
Niedzialkowska, E., Majorek, K.A., Porebski, P.J., Al Obaidi, N., Hammonds, J., Hillerich, B., Seidel, R., Bonanno, J.B., Almo, S.C., Minor, W., New York Structural Genomics Research Consortium (NYSGRC)To be published.