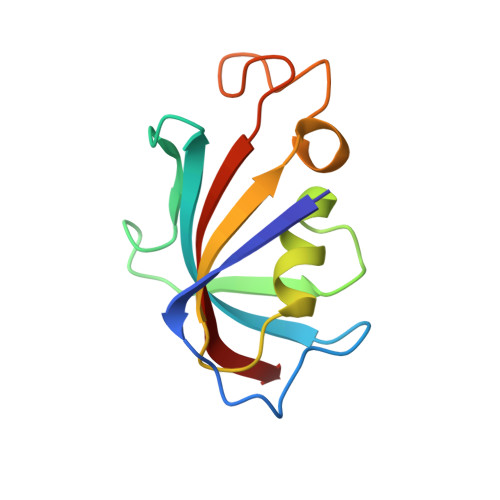
Crystal structure and conformational flexibility of the unligated FK506-binding protein FKBP12.6.
Chen, H., Mustafi, S.M., LeMaster, D.M., Li, Z., Heroux, A., Li, H., Hernandez, G.(2014) Acta Crystallogr D Biol Crystallogr 70: 636-646
- PubMed: 24598733
- DOI: https://doi.org/10.1107/S1399004713032112
- Primary Citation of Related Structures:
4IQ2, 4IQC - PubMed Abstract:
The primary known physiological function of FKBP12.6 involves its role in regulating the RyR2 isoform of ryanodine receptor Ca(2+) channels in cardiac muscle, pancreatic β islets and the central nervous system. With only a single previously reported X-ray structure of FKBP12.6, bound to the immunosuppressant rapamycin, structural inferences for this protein have been drawn from the more extensive studies of the homologous FKBP12. X-ray structures at 1.70 and 1.90 Å resolution from P2₁ and P3₁21 crystal forms are reported for an unligated cysteine-free variant of FKBP12.6 which exhibit a notable diversity of conformations. In one monomer from the P3₁21 crystal form, the aromatic ring of Phe59 at the base of the active site is rotated perpendicular to its typical orientation, generating a steric conflict for the immunosuppressant-binding mode. The peptide unit linking Gly89 and Val90 at the tip of the protein-recognition `80s loop' is flipped in the P2₁ crystal form. Unlike the >30 reported FKBP12 structures, the backbone conformation of this loop closely follows that of the first FKBP domain of FKBP51. The NMR resonances for 21 backbone amides of FKBP12.6 are doubled, corresponding to a slow conformational transition centered near the tip of the 80s loop, as recently reported for 31 amides of FKBP12. The comparative absence of doubling for residues along the opposite face of the active-site pocket in FKBP12.6 may in part reflect attenuated structural coupling owing to increased conformational plasticity around the Phe59 ring.
- Wadsworth Center, New York State Department of Health, Empire State Plaza, Albany, NY 12201, USA.
Organizational Affiliation: