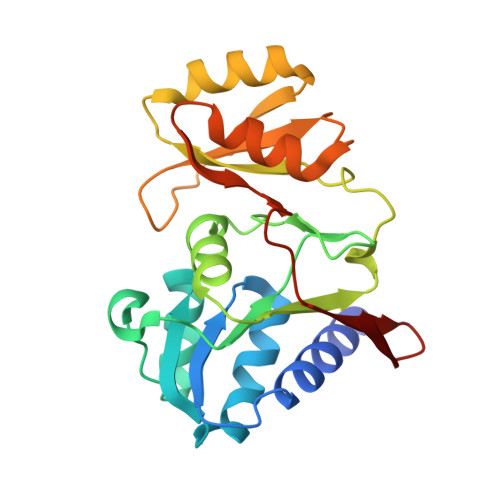
Structural and biophysical studies of Ribose-5-Phosphate Isomerase A from Francisella Tularensis
Rostankowski, R., Orlikowska, M., Nakka, C., Grimshaw, S., Borek, D., Otwinowski, Z., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.