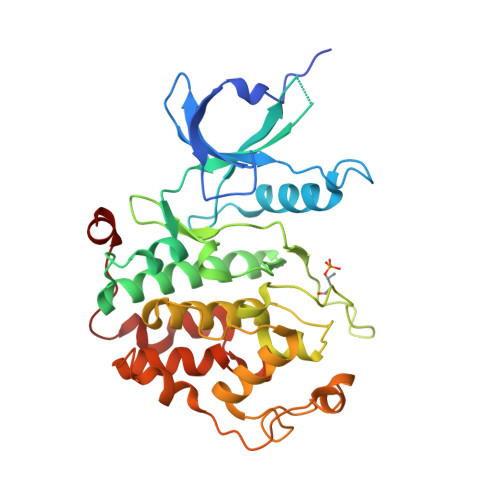
The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat.
Schulze-Gahmen, U., Upton, H., Birnberg, A., Bao, K., Chou, S., Krogan, N.J., Zhou, Q., Alber, T.(2013) Elife 2: e00327-e00327
- PubMed: 23471103
- DOI: https://doi.org/10.7554/eLife.00327
- Primary Citation of Related Structures:
4IMY - PubMed Abstract:
Human positive transcription elongation factor b (P-TEFb) phosphorylates RNA polymerase II and regulatory proteins to trigger elongation of many gene transcripts. The HIV-1 Tat protein selectively recruits P-TEFb as part of a super elongation complex (SEC) organized on a flexible AFF1 or AFF4 scaffold. To understand this specificity and determine if scaffold binding alters P-TEFb conformation, we determined the structure of a tripartite complex containing the recognition regions of P-TEFb and AFF4. AFF4 meanders over the surface of the P-TEFb cyclin T1 (CycT1) subunit but makes no stable contacts with the CDK9 kinase subunit. Interface mutations reduced CycT1 binding and AFF4-dependent transcription. AFF4 is positioned to make unexpected direct contacts with HIV Tat, and Tat enhances P-TEFb affinity for AFF4. These studies define the mechanism of scaffold recognition by P-TEFb and reveal an unanticipated intersubunit pocket on the AFF4 SEC that potentially represents a target for therapeutic intervention against HIV/AIDS. DOI:http://dx.doi.org/10.7554/eLife.00327.001.
- Department of Molecular and Cell Biology , University of California, Berkeley , Berkeley , United States.
Organizational Affiliation: