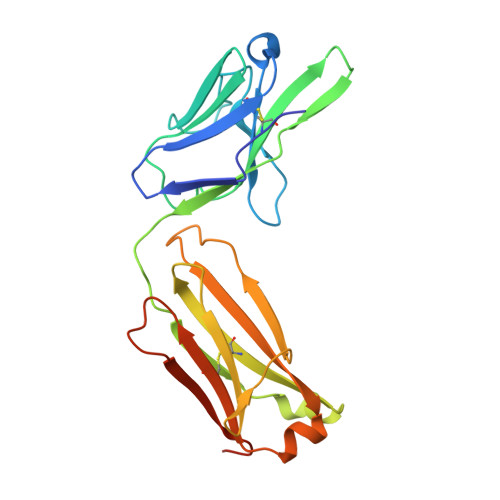
Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Fenn, S., Schiller, C.B., Griese, J.J., Duerr, H., Imhof-Jung, S., Gassner, C., Moelleken, J., Regula, J.T., Schaefer, W., Thomas, M., Klein, C., Hopfner, K.P., Kettenberger, H.(2013) PLoS One 8: e61953-e61953
- PubMed: 23613981
- DOI: https://doi.org/10.1371/journal.pone.0061953
- Primary Citation of Related Structures:
4IMK, 4IML - PubMed Abstract:
Bispecific antibodies are considered as a promising class of future biotherapeutic molecules. They comprise binding specificities for two different antigens, which may provide additive or synergistic modes of action. There is a wide variety of design alternatives for such bispecific antibodies, including the "CrossMab" format. CrossMabs contain a domain crossover in one of the antigen-binding (Fab) parts, together with the "knobs-and-holes" approach, to enforce the correct assembly of four different polypeptide chains into an IgG-like bispecific antibody. We determined the crystal structure of a hAng-2-binding Fab in its crossed and uncrossed form and show that CH1-CL-domain crossover does not induce significant perturbations of the structure and has no detectable influence on target binding.
- Large Molecule Research, Pharma Research and Early Development, Roche Dignostics GmbH, Penzberg, Germany.
Organizational Affiliation: