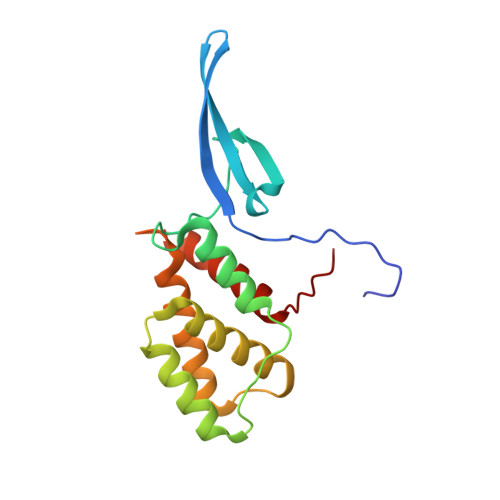
Crystal structure of Mycobacterium tuberculosis CarD, an essential RNA polymerase binding protein, reveals a quasidomain-swapped dimeric structural architecture.
Kaur, G., Dutta, D., Thakur, K.G.(2014) Proteins 82: 879-884
- PubMed: 24115125
- DOI: https://doi.org/10.1002/prot.24419
- Primary Citation of Related Structures:
4ILU, 4MFR - PubMed Abstract:
Mycobacterium tuberculosis (Mtb) CarD is an essential transcriptional regulator that binds RNA polymerase and plays an important role in reprogramming transcription machinery under diverse stress conditions. Here, we report the crystal structure of CarD at 2.3 Å resolution, that represents the first structural description of CarD/CdnL-Like family of proteins. CarD adopts an overall bi-lobed structural architecture where N-terminal domain resembles 'tudor-like' domain and C-terminal domain adopts a novel five helical fold that lacks the predicted leucine zipper structural motif. The structure reveals dimeric state of CarD resulting from β-strand swapping between the N-terminal domains of each individual subunits. The structure provides crucial insights into the possible mode(s) of CarD/RNAP interactions.
- Structural Biology Laboratory, G. N. Ramachandran Protein Centre, CSIR-Institute of Microbial Technology, Chandigarh, 160036, India.
Organizational Affiliation: