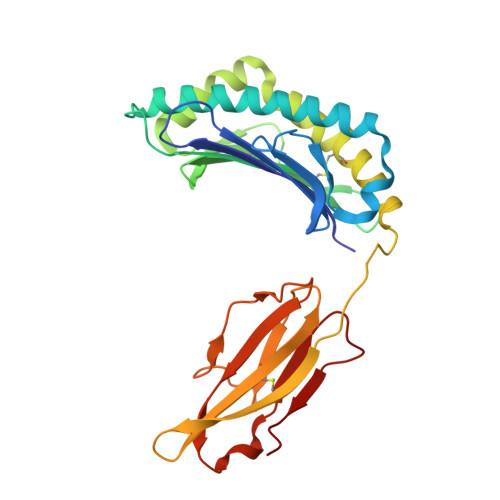
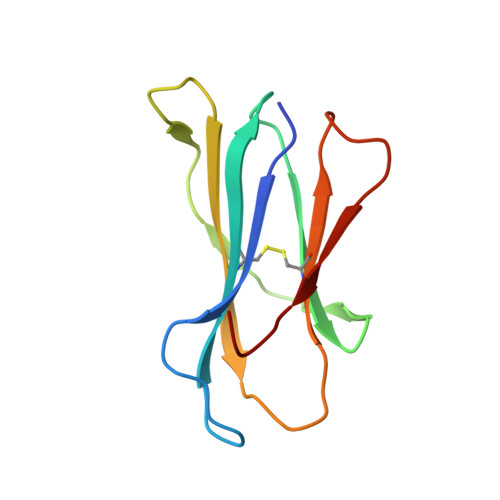
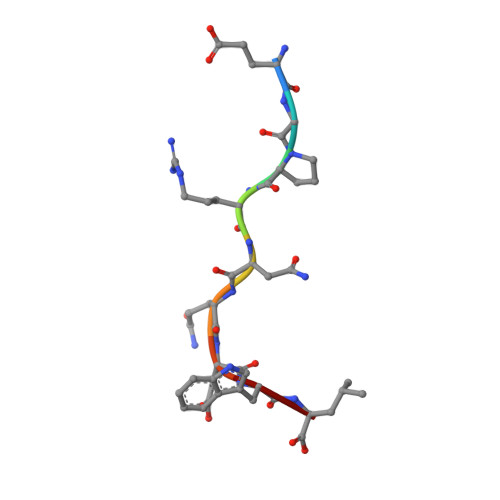
Proline substitution independently enhances H-2D(b) complex stabilization and TCR recognition of melanoma-associated peptides
Uchtenhagen, H., Abualrous, E.T., Stahl, E., Allerbring, E.B., Sluijter, M., Zacharias, M., Sandalova, T., van Hall, T., Springer, S., Nygren, P.A., Achour, A.(2013) Eur J Immunol 43: 3051-3060
- PubMed: 23939911
- DOI: https://doi.org/10.1002/eji.201343456
- Primary Citation of Related Structures:
4IHO - PubMed Abstract:
The immunogenicity of H-2D(b) (D(b)) restricted epitopes can be significantly increased by substituting peptide position 3 to a proline (p3P). The p3P modification enhances MHC stability without altering the conformation of the modified epitope allowing for T-cell cross-reactivity with the native peptide. The present study reveals how specific interactions between p3P and the highly conserved MHC heavy chain residue Y159 increase the stability of D(b) in complex with an optimized version of the melanoma-associated epitope gp10025-33 . Furthermore, the p3P modification directly increased the affinity of the D(b)/gp10025-33 -specific T-cell receptor (TCR) pMel. Surprisingly, the enhanced TCR binding was independent from the observed increased stability of the optimized D(b)/gp10025-33 complex and from the interactions formed between p3P and Y159, indicating a direct effect of the p3P modification on TCR recognition.
- Science for Life Laboratory, Center for Infectious Medicine (CIM), Department of Medicine, Karolinska Insitutet, Karolinska University Hospital Huddinge, Stockholm, Sweden.
Organizational Affiliation: