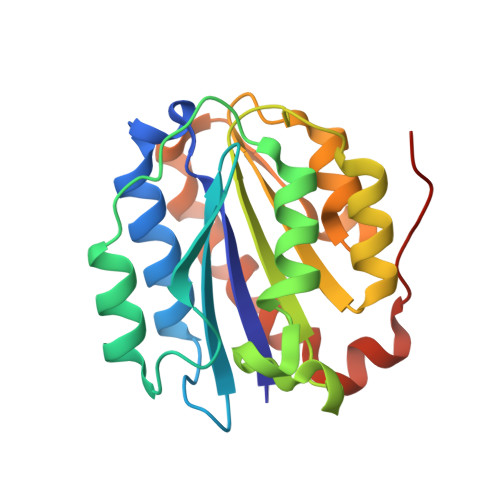
A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein
Becker, A.K., Mikolajek, H., Paulsson, M., Wagener, R., Werner, J.M.(2014) Structure 22: 199-208
- PubMed: 24332716
- DOI: https://doi.org/10.1016/j.str.2013.06.028
- Primary Citation of Related Structures:
4IGI, 4IHK - PubMed Abstract:
Von Willebrand factor A (VWA) domains are versatile protein interaction domains with N and C termini in close proximity placing spatial constraints on overall protein structure. The 1.2 Å crystal structures of a collagen VI VWA domain and a disease-causing point mutant show C-terminal extensions that place the N and C termini at opposite ends. This allows a "beads-on-a-string" arrangement of multiple VWA domains as observed for ten N-terminal domains of the collagen VI α3 chain. The extension is linked to the core domain by a salt bridge and two hydrophobic patches. Comparison of the wild-type and a muscular dystrophy-associated mutant structure identifies a potential perturbation of a protein interaction interface and indeed, the secretion of mutant collagen VI tetramers is affected. Homology modeling is used to locate a number of disease-associated mutations and analyze their structural impact, which will allow mechanistic analysis of collagen-VI-associated muscular dystrophy phenotypes.
- Center for Biochemistry, Medical Faculty, Center for Molecular Medicine Cologne (CMMC), Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne 50931, Germany.
Organizational Affiliation: