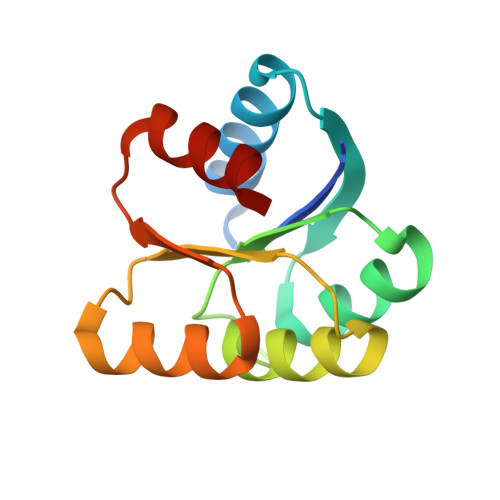
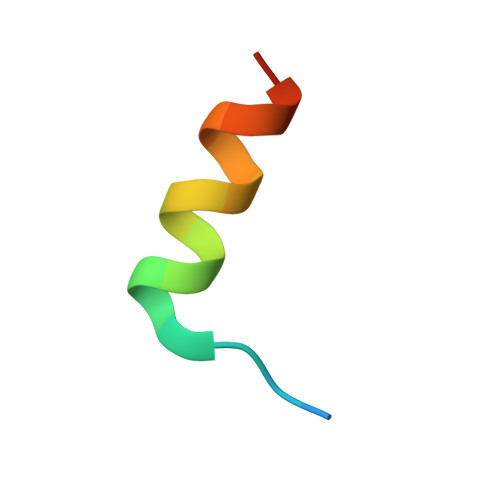
The crystal structure of an activated Thermotoga maritima CheY with N-terminal region of FliM.
Ahn, D.R., Song, H., Kim, J., Lee, S., Park, S.Y.(2013) Int J Biol Macromol 54: 76-83
- PubMed: 23237794
- DOI: https://doi.org/10.1016/j.ijbiomac.2012.12.003
- Primary Citation of Related Structures:
4IGA - PubMed Abstract:
In bacterial chemotaxis, the levels of phosphorylated CheY in association with FliM determine the sense of the flagella rotation, which in turn controls the bacterial swimming behavior. We report the 1.7Å resolution crystallographic structure of the Thermotoga maritima BeF(3)(-)-activated CheY in complex with the CheY-binding N-terminal region of FliM. Analysis of the structure in comparison to the previously reported Escherichia coli counterpart reveals that similar regions of H4-β5-H5 in CheY and the helix in FliM are used for the complex interfaces. Our structure also indicates that the correlated movement of Phe101 and Ser82 (F-S coupling) in T. maritima CheY upon phosphorylation and FliM binding, parallels that of Tyr106 and Thr87 (Y-T coupling) demonstrated in E. coli CheY. Furthermore, significant displacements of the β4-H4 loop in both CheYs impose a crucial role of this loop, which can be related to flagellar switch component binding or to propagating changes that is necessary during the CheY-mediated reversal of the motor.
- Center for Theragnosis, Korea Institute of Science and Technology, Seoul 136-791, Republic of Korea.
Organizational Affiliation: