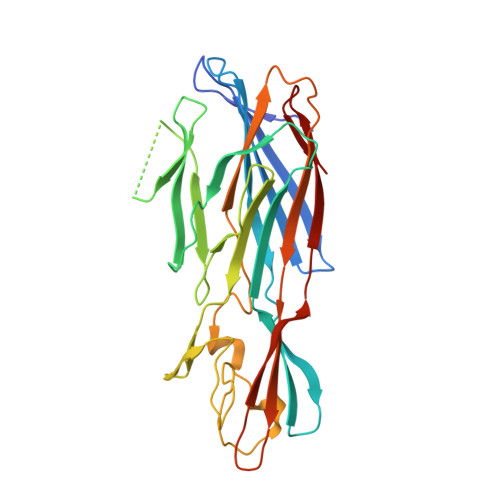
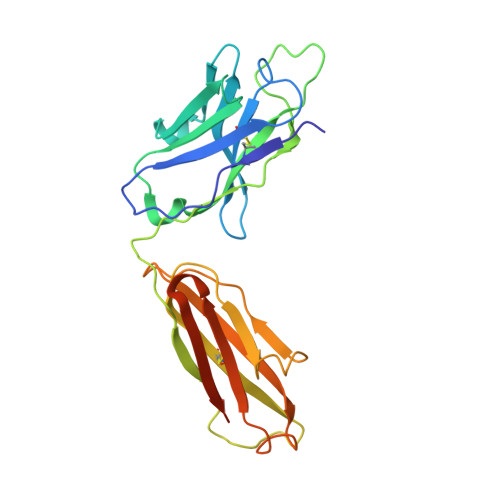
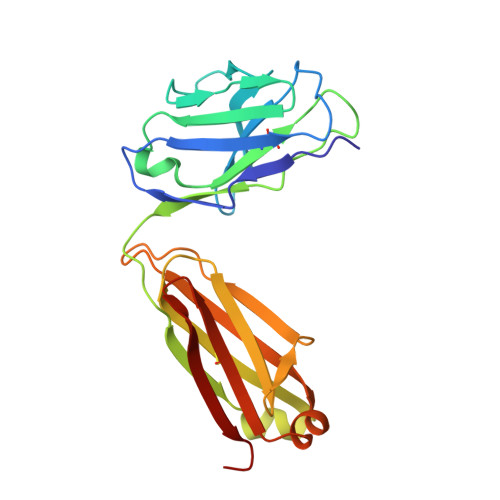
Mechanism of Action and In Vivo Efficacy of a Human-Derived Antibody against Staphylococcus aureus alpha-Hemolysin.
Foletti, D., Strop, P., Shaughnessy, L., Hasa-Moreno, A., Casas, M.G., Russell, M., Bee, C., Wu, S., Pham, A., Zeng, Z., Pons, J., Rajpal, A., Shelton, D.(2013) J Mol Biology 425: 1641-1654
- PubMed: 23416200
- DOI: https://doi.org/10.1016/j.jmb.2013.02.008
- Primary Citation of Related Structures:
4IDJ - PubMed Abstract:
The emergence and spread of multi-drug-resistant strains of Staphylococcus aureus in hospitals and in the community emphasize the urgency for the development of novel therapeutic interventions. Our approach was to evaluate the potential of harnessing the human immune system to guide the development of novel therapeutics. We explored the role of preexisting antibodies against S. aureus α-hemolysin in the serum of human individuals by isolating and characterizing one antibody with a remarkably high affinity to α-hemolysin. The antibody provided protection in S. aureus pneumonia, skin, and bacteremia mouse models of infection and also showed therapeutic efficacy when dosed up to 18 h post-infection in the pneumonia model. Additionally, in pneumonia and bacteremia animal models, the therapeutic efficacy of the α-hemolysin antibody appeared additive to the antibiotic linezolid. To better understand the mechanism of action of this isolated antibody, we solved the crystal structure of the α-hemolysin:antibody complex. To our knowledge, this is the first report of the crystal structure of the α-hemolysin monomer. The structure of the complex shows that the antibody binds α-hemolysin between the cap and the rim domains. In combination with biochemical data, the structure suggests that the antibody neutralizes the activity of the toxin by preventing binding to the plasma membrane of susceptible host cells. The data presented here suggest that protective antibodies directed against S. aureus molecules exist in some individuals and that such antibodies have a therapeutic potential either alone or in combination with antibiotics.
- Rinat Laboratories, Pfizer Inc., 230 East Grand Avenue, South San Francisco, CA 94080, USA. davide.foletti@pfizer.com
Organizational Affiliation: