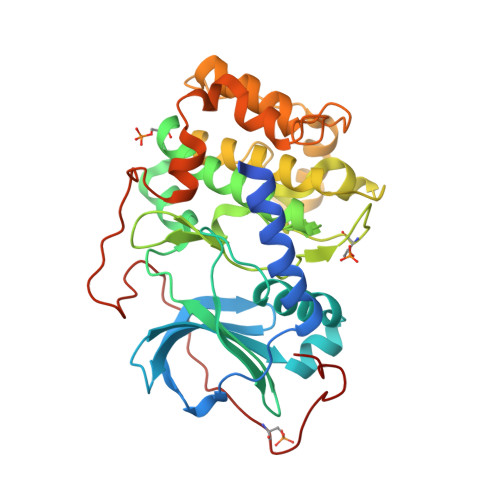
Insights into the Phosphoryl Transfer Catalyzed by cAMP-Dependent Protein Kinase: An X-ray Crystallographic Study of Complexes with Various Metals and Peptide Substrate SP20.
Gerlits, O., Waltman, M.J., Taylor, S., Langan, P., Kovalevsky, A.(2013) Biochemistry 52: 3721-3727
- PubMed: 23672593
- DOI: https://doi.org/10.1021/bi400066a
- Primary Citation of Related Structures:
4IAC, 4IAD, 4IAF, 4IAI, 4IAK, 4IAY, 4IAZ - PubMed Abstract:
X-ray structures of several ternary substrate and product complexes of the catalytic subunit of cAMP-dependent protein kinase (PKAc) have been determined with different bound metal ions. In the PKAc complexes, Mg(2+), Ca(2+), Sr(2+), and Ba(2+) metal ions could bind to the active site and facilitate the phosphoryl transfer reaction. ATP and a substrate peptide (SP20) were modified, and the reaction products ADP and the phosphorylated peptide were found trapped in the enzyme active site. Finally, we determined the structure of a pseudo-Michaelis complex containing Mg(2+), nonhydrolyzable AMP-PCP (β,γ-methyleneadenosine 5'-triphosphate) and SP20. The product structures together with the pseudo-Michaelis complex provide snapshots of different stages of the phosphorylation reaction. Comparison of these structures reveals conformational, coordination, and hydrogen bonding changes that might occur during the reaction and shed new light on its mechanism, roles of metals, and active site residues.
- Bioscience Division, Los Alamos National Laboratory , Los Alamos, New Mexico 87544, United States.
Organizational Affiliation: