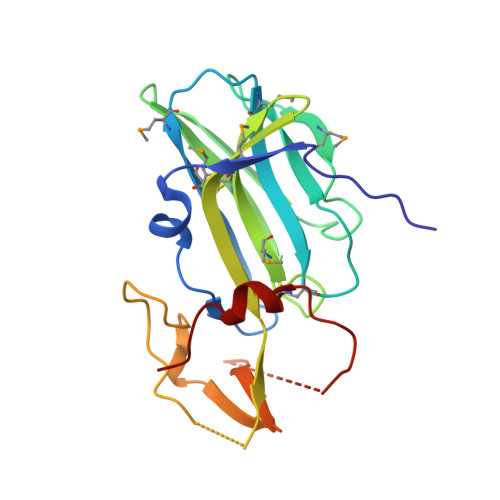
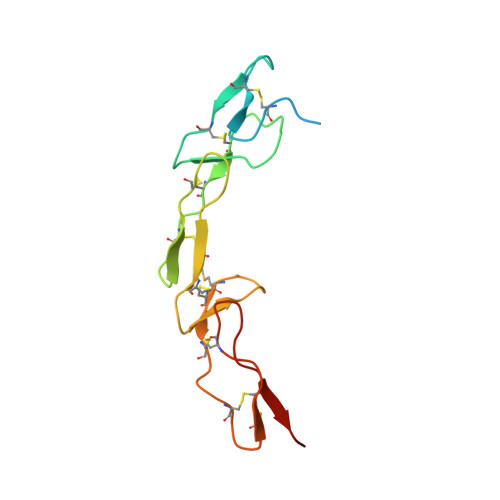
Structure of Human Cytomegalovirus UL141 Binding to TRAIL-R2 Reveals Novel, Non-canonical Death Receptor Interactions.
Nemcovicova, I., Benedict, C.A., Zajonc, D.M.(2013) PLoS Pathog 9: e1003224-e1003224
- PubMed: 23555243
- DOI: https://doi.org/10.1371/journal.ppat.1003224
- Primary Citation of Related Structures:
4I9X - PubMed Abstract:
The TRAIL (TNF-related apoptosis inducing ligand) death receptors (DRs) of the tumor necrosis factor receptor superfamily (TNFRSF) can promote apoptosis and regulate antiviral immunity by maintaining immune homeostasis during infection. In turn, human cytomegalovirus (HCMV) expresses immunomodulatory proteins that down-regulate cell surface expression of TNFRSF members as well as poliovirus receptor-related proteins in an effort to inhibit host immune effector pathways that would lead to viral clearance. The UL141 glycoprotein of human cytomegalovirus inhibits host defenses by blocking cell surface expression of TRAIL DRs (by retention in ER) and poliovirus receptor CD155, a nectin-like Ig-fold molecule. Here we show that the immunomodulatory function of HCMV UL141 is associated with its ability to bind diverse proteins, while utilizing at least two distinct binding sites to selectively engage TRAIL DRs or CD155. Binding studies revealed high affinity interaction of UL141 with both TRAIL-R2 and CD155 and low affinity binding to TRAIL-R1. We determined the crystal structure of UL141 bound to TRAIL-R2 at 2.1 Å resolution, which revealed that UL141 forms a homodimer that engages two TRAIL-R2 monomers 90° apart to form a heterotetrameric complex. Our structural and biochemical data reveal that UL141 utilizes its Ig-domain to facilitate non-canonical death receptor interactions while UL141 partially mimics the binding site of TRAIL on TRAIL-R2, which we found to be distinct from that of CD155. Moreover, UL141 also binds to an additional surface patch on TRAIL-R2 that is distinct from the TRAIL binding site. Therefore, the breadth of UL141-mediated effects indicates that HCMV has evolved sophisticated strategies to evade the immune system by modulating multiple effector pathways.
- Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, California, United States of America.
Organizational Affiliation: