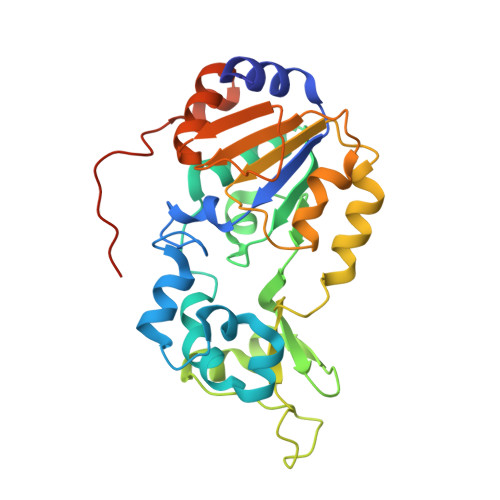
The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
Zhao, X., Allison, D., Condon, B., Zhang, F., Gheyi, T., Zhang, A., Ashok, S., Russell, M., MacEwan, I., Qian, Y., Jamison, J.A., Luz, J.G.(2013) J Med Chem 56: 963-969
- PubMed: 23311358
- DOI: https://doi.org/10.1021/jm301431y
- Primary Citation of Related Structures:
4I5I - PubMed Abstract:
The sirtuin SIRT1 is a NAD(+)-dependent histone deacetylase, a Sir2 family member, and one of seven human sirtuins. Sirtuins are conserved from archaea to mammals and regulate transcription, genome stability, longevity, and metabolism. SIRT1 regulates transcription via deacetylation of transcription factors such as PPARγ, NFκB, and the tumor suppressor protein p53. EX527 (27) is a nanomolar SIRT1 inhibitor and a micromolar SIRT2 inhibitor. To elucidate the mechanism of SIRT inhibition by 27, we determined the 2.5 Å crystal structure of the SIRT1 catalytic domain (residues 241-516) bound to NAD(+) and the 27 analogue compound 35. 35 binds deep in the catalytic cleft, displacing the NAD(+) nicotinamide and forcing the cofactor into an extended conformation. The extended NAD(+) conformation sterically prevents substrate binding. The SIRT1/NAD(+)/35 crystal structure defines a novel mechanism of histone deacetylase inhibition and provides a basis for understanding, and rationally improving, inhibition of this therapeutically important target by drug-like molecules.
- Lilly Biotechnology Center San Diego, 10300 Campus Point Drive, Suite 200, San Diego, California 92121, USA.
Organizational Affiliation: