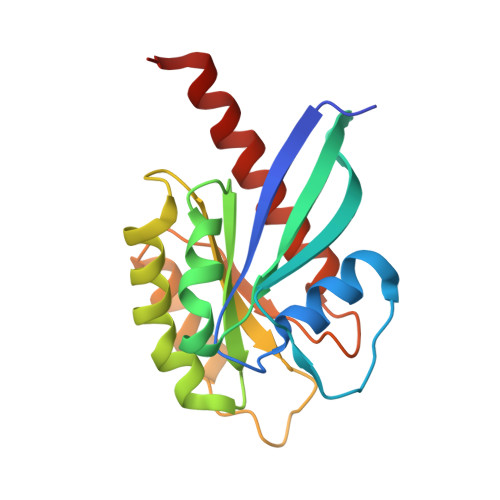
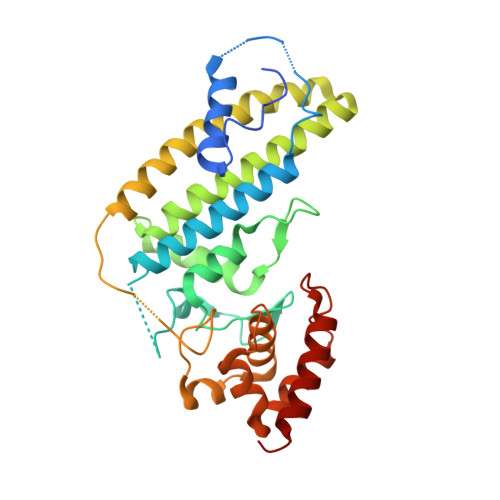
Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB.
Mihai Gazdag, E., Streller, A., Haneburger, I., Hilbi, H., Vetter, I.R., Goody, R.S., Itzen, A.(2013) EMBO Rep 14: 199-205
- PubMed: 23288104
- DOI: https://doi.org/10.1038/embor.2012.211
- Primary Citation of Related Structures:
4I1M, 4I1O - PubMed Abstract:
Legionella pneumophila is an intracellularly surviving pathogen that releases about 270 different proteins into the host cell during infection. A set of secreted proteins takes control of the vesicular trafficking regulator Rab1. Legionella LepB inactivates Rab1 by acting as a GTPase-activating protein (GAP). We present the crystal structure of the Rab1b:LepB complex together with a thorough biochemical analysis and show that the GAP domain of LepB consists of an unusual fold. LepB acts by an atypical RabGAP mechanism that is reminiscent of classical GAPs and therefore sets the protein apart from mammalian TBC-like GAPs. Surprisingly, LepB can function as a GAP for Rab3, Rab8, Rab13 and Rab35, too, suggesting that it has a broader cellular role than previously thought.
- Max-Planck-Institute of Molecular Physiology, Department of Physical Biochemistry, Dortmund 44227, Germany.
Organizational Affiliation: