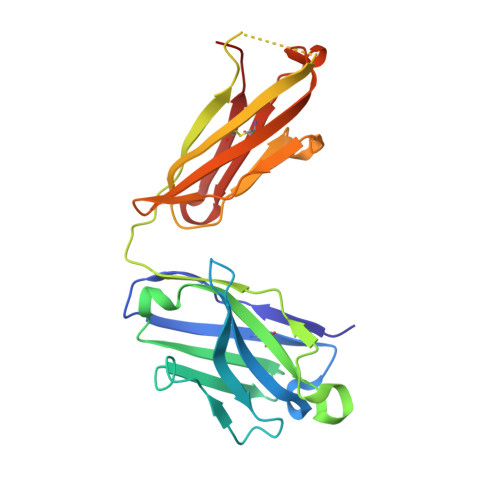
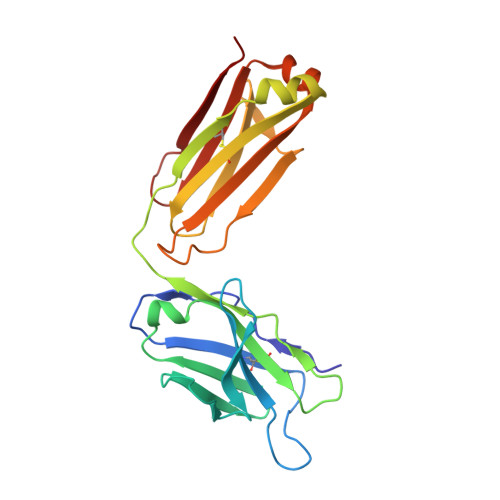
Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus.
Deng, L., Zhong, L., Struble, E., Duan, H., Ma, L., Harman, C., Yan, H., Virata-Theimer, M.L., Zhao, Z., Feinstone, S., Alter, H., Zhang, P.(2013) Proc Natl Acad Sci U S A 110: 7418-7422
- PubMed: 23589879
- DOI: https://doi.org/10.1073/pnas.1305306110
- Primary Citation of Related Structures:
4HZL - PubMed Abstract:
Hepatitis C virus (HCV) envelope glycoprotein E2 has been considered as a major target for vaccine design. Epitope II, mapped between residues 427-446 within the E2 protein, elicits antibodies that are either neutralizing or nonneutralizing. The fundamental mechanism of antibody-mediated neutralization at epitope II remains to be defined at the atomic level. Here we report the crystal structure of the epitope II peptide in complex with a monoclonal antibody (mAb#8) capable of neutralizing HCV. The complex structure revealed that this neutralizing antibody engages epitope II via interactions with both the C-terminal α-helix and the N-terminal loop using a bifurcated mode of action. Our structural insights into the key determinants for the antibody-mediated neutralization may contribute to the immune prophylaxis of HCV infection and the development of an effective HCV vaccine.
- Division of Hematology, Office of Blood Research and Review, Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, MD 20892, USA.
Organizational Affiliation: