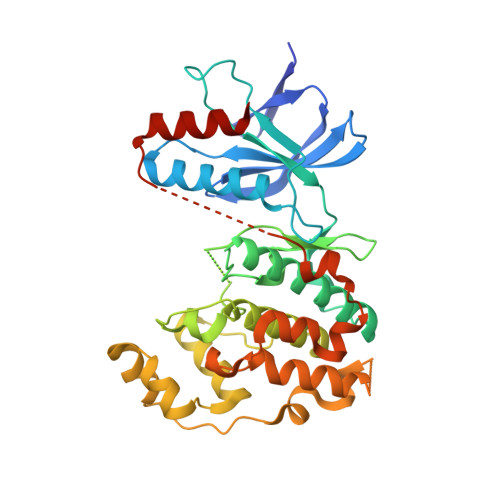
Development of amino-pyrimidine inhibitors of c-Jun N-terminal kinase (JNK): kinase profiling guided optimization of a 1,2,3-benzotriazole lead.
Palmer, W.S., Alam, M., Arzeno, H.B., Chang, K.C., Dunn, J.P., Goldstein, D.M., Gong, L., Goyal, B., Hermann, J.C., Hogg, J.H., Hsieh, G., Jahangir, A., Janson, C., Jin, S., Ursula Kammlott, R., Kuglstatter, A., Lukacs, C., Michoud, C., Niu, L., Reuter, D.C., Shao, A., Silva, T., Trejo-Martin, T.A., Stein, K., Tan, Y.C., Tivitmahaisoon, P., Tran, P., Wagner, P., Weller, P., Wu, S.Y.(2013) Bioorg Med Chem Lett 23: 1486-1492
- PubMed: 23352510
- DOI: https://doi.org/10.1016/j.bmcl.2012.12.047
- Primary Citation of Related Structures:
4HYS, 4HYU - PubMed Abstract:
A series of amino-pyrimidines was developed based upon an initial kinase cross-screening hit from a CDK2 program. Kinase profiling and structure-based drug design guided the optimization from the initial 1,2,3-benzotriazole hit to a potent and selective JNK inhibitor, compound 24f (JNK1 and 2 IC(50)=16 and 66 nM, respectively), with bioavailability in rats and suitable for further in vivo pharmacological evaluation.
- Roche Palo Alto, 3431 Hillview Ave., Palo Alto, CA 94304, USA. wyliepalmer@msn.com
Organizational Affiliation: