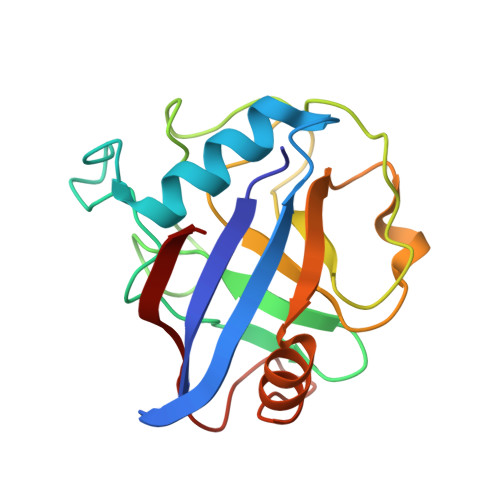
Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1.
Sekhon, S.S., Kaur, H., Dutta, T., Singh, K., Kumari, S., Kang, S., Park, S.G., Park, B.C., Jeong, D.G., Pareek, A., Woo, E.J., Singh, P., Yoon, T.S.(2013) Acta Crystallogr D Biol Crystallogr 69: 555-563
- PubMed: 23519664
- DOI: https://doi.org/10.1107/S0907444912051529
- Primary Citation of Related Structures:
4E1Q, 4HY7 - PubMed Abstract:
Cyclophilins belong to a family of proteins that bind to the immunosuppressive drug cyclosporin A (CsA). Several members of this protein family catalyze the cis-trans isomerization of peptide bonds preceding prolyl residues. The present study describes the biochemical and structural characteristics of a cytosolic cyclophilin (TaCypA-1) cloned from wheat (Triticum aestivum L.). Purified TaCypA-1 expressed in Escherichia coli showed peptidyl-prolyl cis-trans isomerase activity, which was inhibited by CsA with an inhibition constant of 78.3 nM. The specific activity and catalytic efficiency (kcat/Km) of the purified TaCypA-1 were 99.06 ± 0.13 nmol s(-1) mg(-1) and 2.32 × 10(5) M(-1) s(-1), respectively. The structures of apo TaCypA-1 and the TaCypA-1-CsA complex were determined at 1.25 and 1.20 Å resolution, respectively, using X-ray diffraction. Binding of CsA to the active site of TaCypA-1 did not result in any significant conformational change in the apo TaCypA-1 structure. This is consistent with the crystal structure of the human cyclophilin D-CsA complex reported at 0.96 Å resolution. The TaCypA-1 structure revealed the presence of a divergent loop of seven amino acids (48)KSGKPLH(54) which is a characteristic feature of plant cyclophilins. This study is the first to elucidate the structure of an enzymatically active plant cyclophilin which shows peptidyl-prolyl cis-trans isomerase activity and the presence of a divergent loop.
- Medical Proteomics Research Center, Korea Research Institute of Bioscience and Biotechnology, Yuseong-gu, Daejeon, Republic of Korea.
Organizational Affiliation: