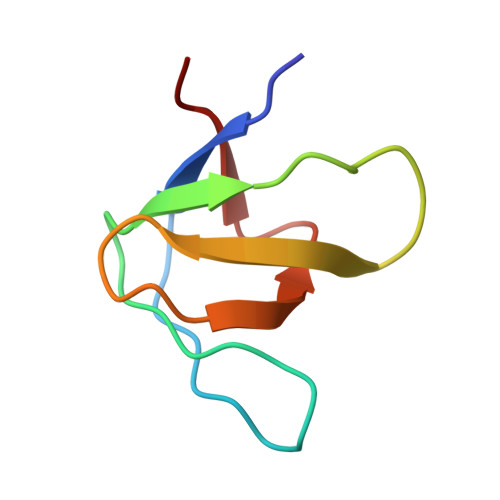
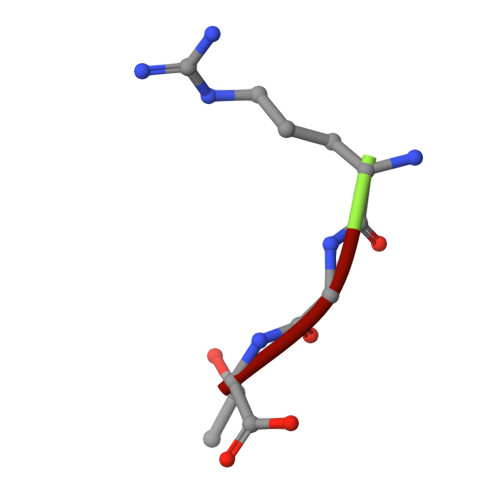
Structural framework of c-Src activation by integrin beta 3
Xiao, R., Xi, X.D., Chen, Z., Chen, S.J., Meng, G.(2013) Blood 121: 700-706
- PubMed: 23169783
- DOI: https://doi.org/10.1182/blood-2012-07-440644
- Primary Citation of Related Structures:
4HXJ - PubMed Abstract:
The integrin β3-mediated c-Src priming and activation, via the SH3 domain, is consistently associated with diseases, such as the formation of thrombosis and the migration of tumor cells. Conventionally, activation of c-Src is often induced by the binding of proline-rich sequences to its SH3 domain. Instead, integrin β3 uses R(760)GT(762) for priming and activation. Because of the lack of structural information, it is not clear where RGT will bind to SH3, and under what mechanism this interaction can prime/activate c-Src. In this study, we present a 2.0-Å x-ray crystal structure in which SH3 is complexed with the RGT peptide. The binding site lies in the "N"-Src loop of the SH3 domain. Structure-based site-directed mutagenesis showed that perturbation on the "N"-Src loop disrupts the interaction between the SH3 domain and the RGT peptide. Furthermore, the simulated c-Src:β3 complex based on the crystal structure of SH3:RGT suggests that the binding of the RGT peptide might disrupt the intramolecular interaction between the SH3 and linker domains, leading to the disengagement of Trp260:"C"-helix and further activation of c-Src.
- State Key Laboratory for Medical Genomics, Shanghai Institute of Hematology, Rui Jin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, People's Republic of China.
Organizational Affiliation: