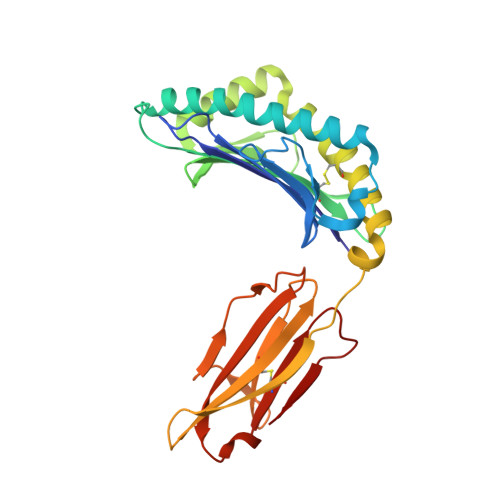
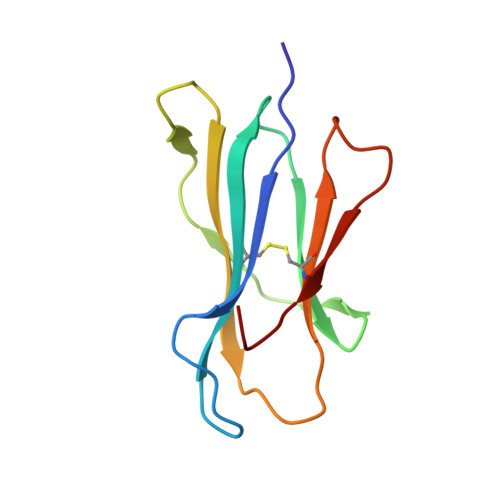
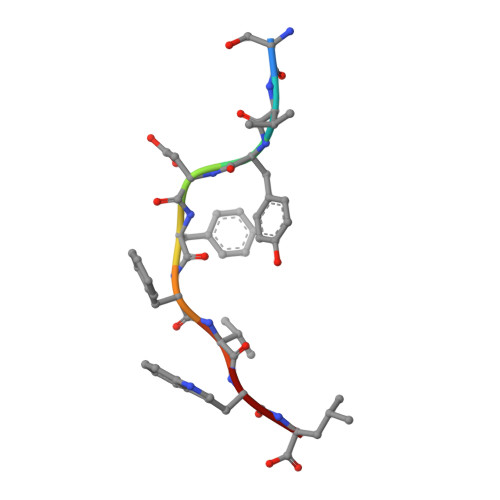
Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Niu, L., Cheng, H., Zhang, S., Tan, S., Zhang, Y., Qi, J., Liu, J., Gao, G.F.(2013) Mol Immunol 55: 381-392
- PubMed: 23566939
- DOI: https://doi.org/10.1016/j.molimm.2013.03.015
- Primary Citation of Related Structures:
4HWZ, 4HX1, 4I48 - PubMed Abstract:
High polymorphism is one of the most important features of human leukocyte antigen (HLA) alleles, which were initially classified by serotyping but have recently been re-grouped into supertypes according to their peptide presentation properties. Two relatively prevalent HLA alleles HLA-A*6801 and HLA-A*6802, are classified into the same serotype HLA-A68. However, based on their distinct peptide-binding characteristics, HLA-A*6801 is grouped into A3 supertype, whereas HLA-A*6802 belongs to A2 supertype, similar to HLA-A*0201. Thusfar, the structural basis of the different supertype definitions of these serotyping-identical HLA alleles remains largely unknown. Herein, we determined the structures of HLA-A*6801 and HLA-A*6802 presenting three typical A3 and A2 supertype-restricted peptides, respectively. The binding capabilities of these peptides to HLA-A*6801, HLA-A*6802, and HLA-A*0201 were analyzed. These data indicate that the similar conformations of the residues within the F pocket contribute to close-related peptide binding features of HLA-A*6802 and HLA-A*0201. However, the overall structure and the peptide conformation of HLA-A*6802 are more similar to HLA-A*6801 rather than HLA-A*0201 which illuminates the similar serotype grouping of HLA-A*6802 and HLA-A*6801. Our findings are helpful for understanding the divergent peptide presentation and virus-specific CTL responses impacted by MHC micropolymorphisms and also elucidate the molecular basis of HLA supertype definitions.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: