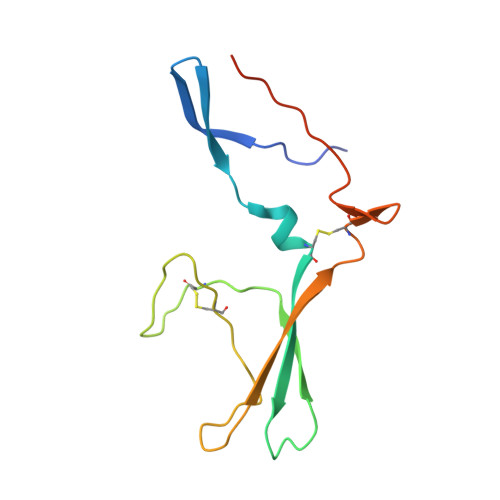
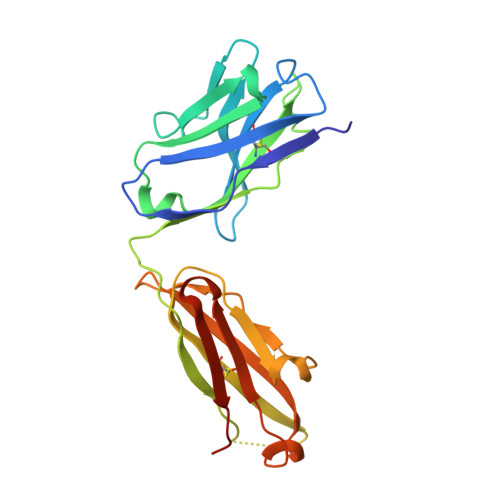
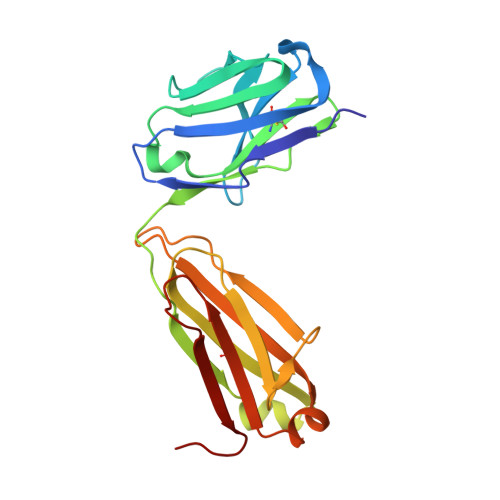
Crystal structure of ectodomain 3 of the IL-13 receptor alpha 1 in complex with a human neutralizing monoclonal antibody fragment
Redpath, N.T., Xu, Y., Wilson, N.J., Fabri, L.J., Baca, M., Andrews, A.E., Braley, H., Lu, P., Ireland, C., Ernst, R.E., Woods, A., Forrest, G., An, Z., Zaller, D.M., Strohl, W.R., Luo, C.S., Czabotar, P.E., Garrett, T.P., Hilton, D.J., Nash, A.D., Zhang, J.G., Nicola, N.A.(2013) Biochem J 451: 165-175
- PubMed: 23384096
- DOI: https://doi.org/10.1042/BJ20121819
- Primary Citation of Related Structures:
4HWB, 4HWE - PubMed Abstract:
Gene deletion studies in mice have revealed critical roles for IL (interleukin)-4 and -13 in asthma development, with the latter controlling lung airways resistance and mucus secretion. We have now developed human neutralizing monoclonal antibodies against human IL-13Rα1 (IL-13 receptor α1) subunit that prevent activation of the receptor complex by both IL-4 and IL-13. We describe the crystal structures of the Fab fragment of antibody 10G5H6 alone and in complex with D3 (ectodomain 3) of IL-13Rα1. Although the structure showed significant domain swapping within a D3 dimer, we showed that Arg(230), Phe(233), Tyr(250), Gln(252) and Leu(293) in each D3 monomer and Ser(32), Asn(102) and Trp(103) in 10G5H6 Fab are the key interacting residues at the interface of the 10G5H6 Fab-D3 complex. One of the most striking contacts is the insertion of the ligand-contacting residue Leu(293) of D3 into a deep pocket on the surface of 10G5H6 Fab, and this appears to be a central determinant of the high binding affinity and neutralizing activity of the antibody.
- The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC 3052, Australia.
Organizational Affiliation: